The novel circFKBP8/miR-432-5p/E2F7 cascade functions as a regulatory network in breast cancer
- PMID: 39192374
- PMCID: PMC11348600
- DOI: 10.1186/s41065-024-00331-1
The novel circFKBP8/miR-432-5p/E2F7 cascade functions as a regulatory network in breast cancer
Abstract
Background: Circular RNAs (circRNAs) are capable of affecting breast cancer (BC) development. However, the role and underneath mechanism of circFKBP8 (also known as hsa_circ_0000915) in BC remain largely unknown.
Methods: Expression analyses were performed using quantitative real-time polymerase chain reaction (qRT-PCR), western blot, and immunohistochemistry (IHC) assays. Effects on cell functional phenotypes were determined by assessing cell proliferation, migratory capacity, invasion, and stemness in vitro. The relationship between microRNA (miR)-432-5p and circFKBP8 or E2F transcription factor 7 (E2F7) was examined by RNA pull-down, dual-luciferase reporter, and RNA immunoprecipitation (RIP) assays. Xenograft assays were used to identify the function of circFKBP8 in vivo.
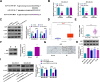
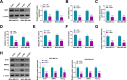
Results: CircFKBP8 was presented at high levels in BC tissues and cells. High circFKBP8 expression was associated with worse overall survival in BC patients. CircFKBP8 suppression inhibited BC cell proliferation, migratory capacity, invasion and stemness in vitro. CircFKBP8 suppression blocked xenograft tumor growth in vivo. Mechanistically, circFKBP8 functioned as a miR-432-5p sponge to modulate E2F7 expression. CircFKBP8 modulated BC cell malignant behaviors by miR-432-5p, and miR-432-5p affected these cell phenotypes through E2F7.
Conclusion: Our observations prove that circFKBP8 promotes BC malignant phenotypes through the miR-432-5p/E2F7 cascade, offering a promising therapeutic and prognostic target for BC.
Keywords: Breast cancer; E2F7; circFKBP8; miR-432-5p.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
-
- Ginsburg O, Bray F, Coleman M, Vanderpuye V, Eniu A, Kotha S, Sarker M, Huong T, Allemani C, Dvaladze A, Gralow J, Yeates K, Taylor C, Oomman N, Krishnan S, Sullivan R, Kombe D, Blas M, Parham G, Kassami N, Conteh L. The global burden of women’s cancers: a grand challenge in global health. Lancet (London England). 2017;389:847–60. 10.1016/s0140-6736(16)31392-7. 10.1016/s0140-6736(16)31392-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

