Similar usage of T-cell receptor β-chain between tumor and adjacent normal tissue in hepatocellular carcinoma
- PMID: 39192502
- PMCID: PMC11349608
- DOI: 10.1002/cam4.70121
Similar usage of T-cell receptor β-chain between tumor and adjacent normal tissue in hepatocellular carcinoma
Abstract
Background: In this study, we comprehensively profiled the T-cell receptor (TCR) repertoire of the tumor and adjacent normal tissue in patients with HBV-associated hepatocellular carcinoma (HCC) and determined the baseline characteristics and clinical significance of TCR.
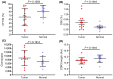
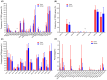
Methods: High-throughput sequencing was used to determine the profile of complementarity-determining region 3 (CDR3) of the TCR-β chain variable (TRBV) in the tumor and normal tissue samples of 14 HCC patients. At the same time, TRBV diversity and differences in expression between tumor and normal tissues were investigated. The cumulative frequency of top 100 CDR3 (CF100), clonality, and Shannon entropy as indices to evaluate diversity, RESULTS: The diversity of TRBV CDR3 showed no significant difference between tumor and normal tissues. Of the 58 V gene segments in TRBV, TRBV16 and TRBV7-6 had a significantly higher frequency in the tumor group than in the normal group (p < 0.05). The frequency of 14 J gene segments showed no significant difference between tumor and normal tissues. In contrast, the frequency of 22 TRBVx/BJx combinations was significantly higher in the tumor than in the normal tissue. In addition, the length and type of TRBV CDR3 were similar in tumor and normal tissues, and a Gaussian distribution was observed in both groups.
Conclusion: This study provided a large amount of information about the TCR lineage in HBV-associated HCC, laying the foundation for further research. In addition, the fact that the immune repertoire (TRBV CDR3) hardly differs between tumor and adjacent normal tissue provides a new clue for exploring the mechanism of the liver as an organ with immune privileges.
Keywords: T‐cell receptor β‐chain; complementarity determining region 3; hepatocellular carcinoma; immune privilege; immune repertoire.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures



References
-
- Pinter M, Scheiner B, Pinato DJ. Immune checkpoint inhibitors in hepatocellular carcinoma: emerging challenges in clinical practice. Lancet Gastroenterol Hepatol. 2023;8:760‐770. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

