The genome sequence of the Small Angle Shades, Euplexia lucipara (Linnaeus, 1758)
- PMID: 39193090
- PMCID: PMC11347913
- DOI: 10.12688/wellcomeopenres.20242.1
The genome sequence of the Small Angle Shades, Euplexia lucipara (Linnaeus, 1758)
Abstract
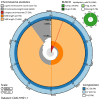
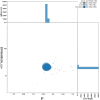
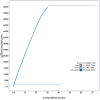
We present a genome assembly from an individual male Euplexia lucipara (the Small Angle Shades; Arthropoda; Insecta; Lepidoptera; Noctuidae). The genome sequence is 661.8 megabases in span. Most of the assembly is scaffolded into 31 chromosomal pseudomolecules, including the Z sex chromosome. The mitochondrial genome has also been assembled and is 15.37 kilobases in length. Gene annotation of this assembly on Ensembl identified 20,395 protein coding genes.
Keywords: Euplexia lucipara; Lepidoptera; Small Angle Shades; chromosomal; genome sequence.
Copyright: © 2023 Boyes D et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


Similar articles
-
The genome sequence of the angle shades moth, Phlogophora meticulosa (Linnaeus, 1758).Wellcome Open Res. 2022 Mar 14;7:89. doi: 10.12688/wellcomeopenres.17757.1. eCollection 2022. Wellcome Open Res. 2022. PMID: 36324701 Free PMC article.
-
The genome sequence of the Burnished Brass, Diachrysia chrysitis (Linnaeus, 1758).Wellcome Open Res. 2023 Feb 15;8:82. doi: 10.12688/wellcomeopenres.18990.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37484482 Free PMC article.
-
The genome sequence of the Dark Spectacle, Abrostola triplasia (Linnaeus, 1758).Wellcome Open Res. 2023 Jun 27;8:278. doi: 10.12688/wellcomeopenres.19624.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 37766851 Free PMC article.
-
The genome sequence of the Hebrew Character, Orthosia gothica (Linnaeus, 1758).Wellcome Open Res. 2024 Feb 19;9:90. doi: 10.12688/wellcomeopenres.20904.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39429633 Free PMC article.
-
The genome sequence of the Heart Moth, Dicycla oo (Linnaeus 1758).Wellcome Open Res. 2023 Jul 21;8:318. doi: 10.12688/wellcomeopenres.19535.1. eCollection 2023. Wellcome Open Res. 2023. PMID: 39092425 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

