Circular RNA IGF1R Promotes Cardiac Repair via Activating β-Catenin Signaling by Interacting with DDX5 in Mice after Ischemic Insults
- PMID: 39193132
- PMCID: PMC11347128
- DOI: 10.34133/research.0451
Circular RNA IGF1R Promotes Cardiac Repair via Activating β-Catenin Signaling by Interacting with DDX5 in Mice after Ischemic Insults
Abstract
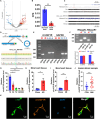
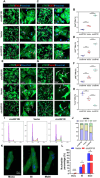
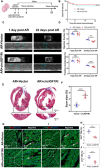
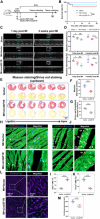
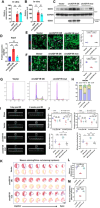
The potential of circular RNAs (circRNAs) as biomarkers and therapeutic targets is becoming increasingly evident, yet their roles in cardiac regeneration and myocardial renewal remain largely unexplored. Here, we investigated the function of circIGF1R and related mechanisms in cardiac regeneration. Through analysis of circRNA sequencing data from neonatal and adult cardiomyocytes, circRNAs associated with regeneration were identified. Our data showed that circIGF1R expression was high in neonatal hearts, decreased with postnatal maturation, and up-regulated after cardiac injury. The elevation was validated in patients diagnosed with acute myocardial infarction (MI) within 1 week. In human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) and myocardial tissue from mice after apical resection and MI, we observed that circIGF1R overexpression enhanced cardiomyocyte proliferation, reduced apoptosis, and mitigated cardiac dysfunction and fibrosis, while circIGF1R knockdown impeded endogenous cardiac renewal. Mechanistically, we identified circIGF1R binding proteins through circRNA precipitation followed by mass spectrometry. RNA pull-down Western blot and RNA immunoprecipitation demonstrated that circIGF1R directly interacted with DDX5 and augmented its protein level by suppressing ubiquitin-dependent degradation. This subsequently triggered the β-catenin signaling pathway, leading to the transcriptional activation of cyclin D1 and c-Myc. The roles of circIGF1R and DDX5 in cardiac regeneration were further substantiated through site-directed mutagenesis and rescue experiments. In conclusion, our study highlights the pivotal role of circIGF1R in facilitating heart regeneration and repair after ischemic insults. The circIGF1R/DDX5/β-catenin axis emerges as a novel therapeutic target for enhancing myocardial repair after MI, offering promising avenues for the development of regenerative therapies.
Copyright © 2024 Tian-Kai Shan et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures



References
-
- Ambrosy AP, Fonarow GC, Butler J, Chioncel O, Greene SJ, Vaduganathan M, Nodari S, Lam CSP, Sato N, Shah AN, et al. The global health and economic burden of hospitalizations for heart failure: Lessons learned from hospitalized heart failure registries. J Am Coll Cardiol. 2014;63(12):1123–1133. - PubMed
-
- Docherty KF, Ferreira JP, Sharma A, Girerd N, Gregson J, Duarte K, Petrie MC, Jhund PS, Dickstein K, Pfeffer MA, et al. Predictors of sudden cardiac death in high-risk patients following a myocardial infarction. Eur J Heart Fail. 2020;22(5):848–855. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

