Exploring protein functions from structural flexibility using CABS-flex modeling
- PMID: 39194135
- PMCID: PMC11350595
- DOI: 10.1002/pro.5090
Exploring protein functions from structural flexibility using CABS-flex modeling
Abstract
Understanding protein function often necessitates characterizing the flexibility of protein structures. However, simulating protein flexibility poses significant challenges due to the complex dynamics of protein systems, requiring extensive computational resources and accurate modeling techniques. In response to these challenges, the CABS-flex method has been developed as an efficient modeling tool that combines coarse-grained simulations with all-atom detail. Available both as a web server and a standalone package, CABS-flex is dedicated to a wide range of users. The web server version offers an accessible interface for straightforward tasks, while the standalone command-line program is designed for advanced users, providing additional features, analytical tools, and support for handling large systems. This paper examines the application of CABS-flex across various structure-function studies, facilitating investigations into the interplay among protein structure, dynamics, and function in diverse research fields. We present an overview of the current status of the CABS-flex methodology, highlighting its recent advancements, practical applications, and forthcoming challenges.
Keywords: Monte Carlo simulation; coarse‐grained model; integrative modeling; molecular modeling; multiscale simulation; protein aggregation; protein flexibility.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


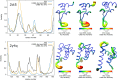

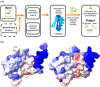

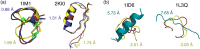

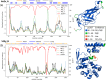
References
-
- Badaczewska‐Dawid AE, Khramushin A, Kolinski A, Schueler‐Furman O, Kmiecik S. Protocols for all‐atom reconstruction and high‐resolution refinement of protein–peptide complex structures. In: Kihara D, editor. Protein structure prediction. Methods in molecular biology. New York, NY: Springer US; 2020a. p. 273–287. 10.1007/978-1-0716-0708-4_16 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

