Requirement of the N-terminal region of nonstructural protein 1 in cis for SARS-CoV-2 defective RNA replication
- PMID: 39194239
- PMCID: PMC11406973
- DOI: 10.1128/jvi.00900-24
Requirement of the N-terminal region of nonstructural protein 1 in cis for SARS-CoV-2 defective RNA replication
Abstract
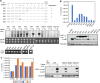
SARS-CoV-2 belongs to the family Coronaviridae and carries a single-stranded positive-sense RNA genome. During coronavirus (CoV) replication, defective or defective interfering RNAs that lack a large portion of the genome often emerge. These defective RNAs typically carry the necessary RNA elements that are required for replication and packaging. We identified the minimum requirement of the 5' proximal region necessary for viral RNA replication by using artificially generated SARS-CoV-2 minigenomes. The minigenomes consist of the 5'-proximal region, an open reading frame (ORF) that encodes a fusion protein consisting of the N-terminal of viral NSP1 and a reporter gene, and the 3' untranslated region of the SARS-CoV-2 genome. We used a modified SARS-CoV-2 variant to support replication of the minigenomes. A minigenome carrying the 5' proximal 634 nucleotides replicated, whereas those carrying shorter than 634 nucleotides did not, demonstrating that the entire 265 nt-long 5' untranslated region and N-terminal portion of the NSP1 coding region are required for the minigenome replication. Minigenome RNAs carrying a specific amino acid substitution or frame shift insertions in the partial NSP1 coding sequence abrogated minigenome replication. Introduction of synonymous mutations in the minigenome RNAs also affected the replication efficiency of the minigenomes. These data suggest that the expression of the N-terminal portion of NSP1 and the primary sequence of the 5' proximal 634 nucleotides are important for minigenome replication.IMPORTANCESARS-CoV-2, the causative agent of COVID-19, is highly transmissible and continues to have a significant impact on public health and the global economy. While several vaccines mitigate the severe consequences of SARS-CoV-2 infection, mutant viruses with reduced reactivity to current vaccines continue to emerge and circulate. This study aimed to identify the minimal 5' proximal region of SARS-CoV-2 genomic RNA required for SARS-CoV-2 defective RNA replication and investigate the importance of an ORF encoded in these defective RNAs. Identifying cis-acting replication signals of SARS-CoV-2 genomic RNA is critical for the development of antivirals that target these signals. Additionally, replication-competent defective RNAs can serve as therapeutic reagents to interfere with SARS-CoV-2 replication. Our findings provide valuable insights into the mechanisms of SARS-CoV-2 RNA replication and the development of reagents that suppress SARS-CoV-2 replication.
Keywords: SARS-CoV-2; cis-acting replication signal; coronavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Dependence of coronavirus RNA replication on an NH2-terminal partial nonstructural protein 1 in cis.J Virol. 2014 Aug;88(16):8868-82. doi: 10.1128/JVI.00738-14. Epub 2014 May 28. J Virol. 2014. PMID: 24872586 Free PMC article.
-
Eight-amino-acid sequence at the N-terminus of SARS-CoV-2 nsp1 is involved in stabilizing viral genome replication.Virology. 2024 Jul;595:110068. doi: 10.1016/j.virol.2024.110068. Epub 2024 Apr 8. Virology. 2024. PMID: 38593595
-
Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements.J Virol. 2009 Jun;83(12):6087-97. doi: 10.1128/JVI.00160-09. Epub 2009 Apr 8. J Virol. 2009. PMID: 19357173 Free PMC article.
-
Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19.Cells. 2021 Apr 6;10(4):821. doi: 10.3390/cells10040821. Cells. 2021. PMID: 33917481 Free PMC article. Review.
-
Replication of the coronavirus genome: A paradox among positive-strand RNA viruses.J Biol Chem. 2022 May;298(5):101923. doi: 10.1016/j.jbc.2022.101923. Epub 2022 Apr 10. J Biol Chem. 2022. PMID: 35413290 Free PMC article. Review.
Cited by
-
Characterization of the binding features between SARS-CoV-2 5'-proximal transcripts of genomic RNA and nucleocapsid proteins.RNA Biol. 2025 Dec;22(1):1-16. doi: 10.1080/15476286.2025.2471643. Epub 2025 Mar 12. RNA Biol. 2025. PMID: 40077853 Free PMC article. Review.
References
-
- Bayani F, Hashkavaei NS, Arjmand S, Rezaei S, Uskoković V, Alijanianzadeh M, Uversky VN, Ranaei Siadat SO, Mozaffari-Jovin S, Sefidbakht Y. 2023. An overview of the vaccine platforms to combat COVID-19 with a focus on the subunit vaccines. Prog Biophys Mol Biol 178:32–49. doi:10.1016/j.pbiomolbio.2023.02.004 - DOI - PMC - PubMed
-
- Hyun HJ, Choi MJ, Nham E, Seong H, Yoon JG, Noh JY, Cheong HJ, Kim WJ, Yoon SK, Park SJ, Gwak WS, Lee JW, Kim BG, Song JY. 2023. Neutralizing activity against BQ.1.1, BN.1, and XBB.1 in bivalent COVID-19 vaccine recipients: comparison by the types of prior infection and vaccine formulations. Vaccines (Basel) 11:1320. doi:10.3390/vaccines11081320 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

