A reporter Oropouche virus expressing ZsGreen from the M segment enables pathogenesis studies in mice
- PMID: 39194249
- PMCID: PMC11406970
- DOI: 10.1128/jvi.00893-24
A reporter Oropouche virus expressing ZsGreen from the M segment enables pathogenesis studies in mice
Abstract
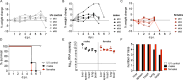
Oropouche fever caused by Oropouche virus (OROV) is a significant zoonosis in Central and South America. Despite its public health significance, we lack high-throughput diagnostics, therapeutics, and a comprehensive knowledge of OROV biology. Reporter viruses are valuable tools to rapidly study virus dynamics and develop neutralization and antiviral screening assays. OROV is a tri-segmented bunyavirus, which makes generating a reporter virus challenging, as introducing foreign elements into the viral genome typically affects fitness. We previously demonstrated that the non-structural gene NSm on the OROV medium (M) segment is non-essential for replication in vitro. Taking advantage of this, we have now generated a recombinant OROV expressing fluorescent protein ZsGreen in place of NSm. This reporter OROV is both stable and pathogenic in IFNAR-/- mice and provides a powerful tool for OROV pathogenesis studies and assay development.IMPORTANCEEmerging and reemerging infectious agents such as zoonotic bunyaviruses are of global health concern. Oropouche virus (OROV) causes recurring outbreaks of acute febrile illness in the Central and South American human populations. Biting midges are the primary transmission vectors, whereas sloths and non-human primates are their reservoir hosts. As global temperatures increase, we will likely see an expansion in arthropod-borne pathogens such as OROV. Therefore, developing reagents to study pathogen biology to aid in identifying druggable targets is essential. Here, we demonstrate the feasibility and use of a fluorescent OROV reporter in mice to study viral dynamics and pathogenesis. We show that this reporter OROV maintains characteristics such as growth and pathogenicity similar to the wild-type virus. Using this reporter virus, we can now develop methods to assist OROV studies and establish various high-throughput assays.
Keywords: Oropouche virus; bunyavirus; fluorescent virus; pathogenesis; reporter virus; tropism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

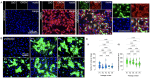
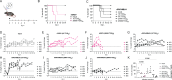

References
-
- Tilston-Lunel NL, Hughes J, Acrani GO, da Silva DEA, Azevedo RSS, Rodrigues SG, Vasconcelos PFC, Nunes MRT, Elliott RM. 2015. Genetic analysis of members of the species Oropouche virus and identification of a novel M segment sequence. J Gen Virol 96:1636–1650. doi:10.1099/vir.0.000108 - DOI - PMC - PubMed
-
- Acrani GO, Tilston-Lunel NL, Spiegel M, Weidmann M, Dilcher M, Andrade da Silva DE, Nunes MRT, Elliott RM. 2015. Establishment of a minigenome system for Oropouche virus reveals the S genome segment to be significantly longer than reported previously. J Gen Virol 96:513–523. doi:10.1099/jgv.0.000005 - DOI - PubMed
-
- Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Ishak R, Freitas RB, Gomes ML, LeDuc JW, Oliva OF. 1981. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am J Trop Med Hyg 30:149–160. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

