Analytical and clinical validation of direct detection of antimicrobial resistance markers by plasma microbial cell-free DNA sequencing
- PMID: 39194269
- PMCID: PMC11481525
- DOI: 10.1128/jcm.00425-24
Analytical and clinical validation of direct detection of antimicrobial resistance markers by plasma microbial cell-free DNA sequencing
Abstract
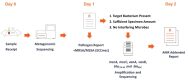
Sequencing of plasma microbial cell-free DNA (mcfDNA) has gained increased acceptance as a valuable adjunct to standard-of-care testing for diagnosis of infections throughout the body. Here, we report the analytical and clinical validation of a novel application of mcfDNA sequencing, the non-invasive detection of seven common antimicrobial resistance (AMR) genetic markers in 18 important pathogens. The AMR markers include SCCmec, mecA, mecC, vanA, vanB, blaCTX-M, and blaKPC. The AMR markers were computationally linked to the pathogens detected. Analytical validation showed high reproducibility (100%), inclusivity (54 to 100%), and exclusivity (100%). Clinical accuracy was assessed with 114 unique plasma samples from patients at seven study sites with concordant culture results for target bacteria from a variety of specimen types and correlated with available phenotypic antimicrobial susceptibility test results and genotypic results. The positive percent agreement (PPA), negative percent agreement (NPA), and diagnostic yield (DY) were estimated for each AMR marker. DY was defined as the percentage of tests that yielded an actionable result of either detected or not detected. The results for the combination of SCCmec and mecA for staphylococci were PPA 19/20 (95.0%), NPA 21/22 (95.4%), DY 42/60 (70.0%); vanA for enterococci were PPA 3/3 (100%), NPA 2/2 (100%), DY 5/6 (83.3%); blaCTX-M for gram-negative bacilli were PPA 5/6 (83.3%), NPA 29/29 (100%), DY 35/49 (71.4%); and blaKPC for gram-negative bacilli were PPA 0/2 (0%), NPA: 23/23 (100%), DY 25/44 (56.8%). The addition of AMR capability to plasma mcfDNA sequencing should provide clinicians with an effective new culture-independent tool for optimization of therapy.
Importance: This manuscript is ideally suited for the Innovative Diagnostic Methods sections as it reports the analytical and clinical validation of a novel application of plasma microbial cell-free DNA sequencing for direct detection of seven selected antimicrobial resistance markers in 18 target pathogens. Clearly, it has potential clinical utility in optimizing therapy and was incorporated into the Karius test workflow in September 2023. In addition, the workflow could readily be adapted to expand the number of target bacteria and antimicrobial resistance markers as needed.
Keywords: antimicrobial resistance genes; metagenomics; microbial cell-free DNA.
Conflict of interest statement
All employees of Karius, which markets the commercial test evaluated in this study, shared responsibilities for study design, analysis of the data, and writing and editing of the manuscript. John Joseph Farrell (OSF Healthcare), Sanjeet Dadwal (City of Hope), Amy L. Carr (AdventHealth Orlando), Arryn Craney (Orlando Health), Matthew Pike (Carle Foundation Hospital), and James B. Wood (Indiana University School of Medicine) received grant support from Karius to support the collection and curation of orthogonal data that were fundamental to the clinical validation and reviewed the manuscript.
Figures
Similar articles
-
Multicenter Evaluation of the Acuitas AMR Gene Panel for Detection of an Extended Panel of Antimicrobial Resistance Genes among Bacterial Isolates.J Clin Microbiol. 2022 Mar 16;60(3):e0209821. doi: 10.1128/JCM.02098-21. Epub 2022 Mar 16. J Clin Microbiol. 2022. PMID: 35138924 Free PMC article.
-
Evaluation and clinical practice of pathogens and antimicrobial resistance genes of BioFire FilmArray Pneumonia panel in lower respiratory tract infections.Infection. 2024 Apr;52(2):545-555. doi: 10.1007/s15010-023-02144-2. Epub 2023 Dec 20. Infection. 2024. PMID: 38123753 Free PMC article.
-
Phenotypic and genotypic perspectives on detection methods for bacterial antimicrobial resistance in a One Health context: research progress and prospects.Arch Microbiol. 2024 Sep 20;206(10):409. doi: 10.1007/s00203-024-04131-z. Arch Microbiol. 2024. PMID: 39302440 Review.
-
MultiSeq-AMR: a modular amplicon-sequencing workflow for rapid detection of bloodstream infection and antimicrobial resistance markers.Microb Genom. 2025 Apr;11(4). doi: 10.1099/mgen.0.001383. Microb Genom. 2025. PMID: 40178523
-
Rapid molecular tests for detection of antimicrobial resistance determinants in Gram-negative organisms from positive blood cultures: a systematic review and meta-analysis.Clin Microbiol Infect. 2020 Mar;26(3):271-280. doi: 10.1016/j.cmi.2019.11.009. Epub 2019 Nov 18. Clin Microbiol Infect. 2020. PMID: 31751768
Cited by
-
Clinical Utility of Plasma Microbial Cell-Free DNA Surveillance in Neutropenic Patients with Acute Myeloid Leukemia Undergoing Outpatient Chemotherapy: A Case Series.Diagnostics (Basel). 2025 Jul 5;15(13):1715. doi: 10.3390/diagnostics15131715. Diagnostics (Basel). 2025. PMID: 40647714 Free PMC article.
References
-
- Mestrovic T, Robles Aguilar G, Swetschinski LR, Ikuta KS, Gray AP, Davis Weaver N, Han C, Wool EE, Gershberg Hayoon A, Hay SI, et al. . 2022. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: a cross-country systematic analysis. Lancet Public Health 7:e897–e913. doi:10.1016/S2468-2667(22)00225-0 - DOI - PMC - PubMed
-
- Uprety P, Kirn TJ. 2024. Chapter 27 - molecular detection of antimicrobial resistance, p 527–531. In Tang YW, Hindiyeh MY, Liu D, Sails A, Spearman P, Zhang JR (ed), Molecular medical microbiology, Third Edition. Academic Press.
-
- Lewinski MA, Alby K, Babady NE, Butler-Wu SM, Bard JD, Greninger AL, Hanson K, Naccache SN, Newton D, Temple-Smolkin RL, Nolte F. 2023. Exploring the utility of multiplex infectious disease panel testing for diagnosis of infection in different body sites: a joint report of the association for molecular pathology, American society for microbiology, infectious diseases society of America, and Pan American society for clinical virology. J Mol Diagn 25:857–875. doi:10.1016/j.jmoldx.2023.08.005 - DOI - PubMed
-
- Webber DM, Wallace MA, Burnham C-A, Anderson NW. 2020. Evaluation of the BioFire FilmArray pneumonia panel for detection of viral and bacterial pathogens in lower respiratory tract specimens in the setting of a tertiary care academic medical center. J Clin Microbiol 58:e00343-20. doi:10.1128/JCM.00343-20 - DOI - PMC - PubMed
-
- Klein M, Bacher J, Barth S, Atrzadeh F, Siebenhaller K, Ferreira I, Beisken S, Posch AE, Carroll KC, Wunderink RG, Qi C, Wu F, Hardy DJ, Patel R, Sims MD. 2021. Multicenter evaluation of the Unyvero platform for testing bronchoalveolar lavage fluid. J Clin Microbiol 59:02497–20. doi:10.1128/JCM.02497-20 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

