Precise Image-level Localization of Intracranial Hemorrhage on Head CT Scans with Deep Learning Models Trained on Study-level Labels
- PMID: 39194400
- PMCID: PMC11605431
- DOI: 10.1148/ryai.230296
Precise Image-level Localization of Intracranial Hemorrhage on Head CT Scans with Deep Learning Models Trained on Study-level Labels
Abstract
Purpose To develop a highly generalizable weakly supervised model to automatically detect and localize image-level intracranial hemorrhage (ICH) by using study-level labels. Materials and Methods In this retrospective study, the proposed model was pretrained on the image-level Radiological Society of North America dataset and fine-tuned on a local dataset by using attention-based bidirectional long short-term memory networks. This local training dataset included 10 699 noncontrast head CT scans in 7469 patients, with ICH study-level labels extracted from radiology reports. Model performance was compared with that of two senior neuroradiologists on 100 random test scans using the McNemar test, and its generalizability was evaluated on an external independent dataset. Results The model achieved a positive predictive value (PPV) of 85.7% (95% CI: 84.0, 87.4) and an area under the receiver operating characteristic curve of 0.96 (95% CI: 0.96, 0.97) on the held-out local test set (n = 7243, 3721 female) and 89.3% (95% CI: 87.8, 90.7) and 0.96 (95% CI: 0.96, 0.97), respectively, on the external test set (n = 491, 178 female). For 100 randomly selected samples, the model achieved performance on par with two neuroradiologists, but with a significantly faster (P < .05) diagnostic time of 5.04 seconds per scan (vs 86 seconds and 22.2 seconds for the two neuroradiologists, respectively). The model's attention weights and heatmaps visually aligned with neuroradiologists' interpretations. Conclusion The proposed model demonstrated high generalizability and high PPVs, offering a valuable tool for expedited ICH detection and prioritization while reducing false-positive interruptions in radiologists' workflows. Keywords: Computer-Aided Diagnosis (CAD), Brain/Brain Stem, Hemorrhage, Convolutional Neural Network (CNN), Transfer Learning Supplemental material is available for this article. © RSNA, 2024 See also the commentary by Akinci D'Antonoli and Rudie in this issue.
Keywords: Brain/Brain Stem; Computer-Aided Diagnosis (CAD); Convolutional Neural Network (CNN); Hemorrhage; Transfer Learning.
Conflict of interest statement
Figures



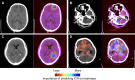
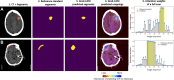
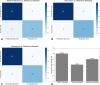
Comment in
-
Achieving More with Less: Combining Strong and Weak Labels for Intracranial Hemorrhage Detection.Radiol Artif Intell. 2024 Nov;6(6):e240670. doi: 10.1148/ryai.240670. Radiol Artif Intell. 2024. PMID: 39503591 Free PMC article. No abstract available.
References
-
- RSNA Intracranial Hemorrhage Detection . https://kaggle.com/competitions/rsna-intracranial-hemorrhage-detection. Accessed October 26, 2021 .
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

