Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies
- PMID: 39194576
- PMCID: PMC11351889
- DOI: 10.3390/biology13080638
Deciphering Depression: Epigenetic Mechanisms and Treatment Strategies
Abstract
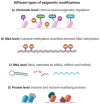
Depression, a significant mental health disorder, is under intense research scrutiny to uncover its molecular foundations. Epigenetics, which focuses on controlling gene expression without altering DNA sequences, offers promising avenues for innovative treatment. This review explores the pivotal role of epigenetics in depression, emphasizing two key aspects: (I) identifying epigenetic targets for new antidepressants and (II) using personalized medicine based on distinct epigenetic profiles, highlighting potential epigenetic focal points such as DNA methylation, histone structure alterations, and non-coding RNA molecules such as miRNAs. Variations in DNA methylation in individuals with depression provide opportunities to target genes that are associated with neuroplasticity and synaptic activity. Aberrant histone acetylation may indicate that antidepressant strategies involve enzyme modifications. Modulating miRNA levels can reshape depression-linked gene expression. The second section discusses personalized medicine based on epigenetic profiles. Analyzing these patterns could identify biomarkers associated with treatment response and susceptibility to depression, facilitating tailored treatments and proactive mental health care. Addressing ethical concerns regarding epigenetic information, such as privacy and stigmatization, is crucial in understanding the biological basis of depression. Therefore, researchers must consider these issues when examining the role of epigenetics in mental health disorders. The importance of epigenetics in depression is a critical aspect of modern medical research. These findings hold great potential for novel antidepressant medications and personalized treatments, which would significantly improve patient outcomes, and transform psychiatry. As research progresses, it is expected to uncover more complex aspects of epigenetic processes associated with depression, enhance our comprehension, and increase the effectiveness of therapies.
Keywords: DNA methylation; antidepressant drugs; biomarkers; epigenetics; histone modifications; non-coding RNAs; personalized medicine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

