Far-Red Fluorescent Proteins: Tools for Advancing In Vivo Imaging
- PMID: 39194588
- PMCID: PMC11352758
- DOI: 10.3390/bios14080359
Far-Red Fluorescent Proteins: Tools for Advancing In Vivo Imaging
Abstract
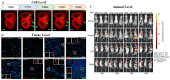
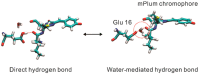
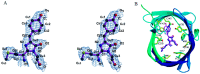
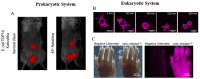
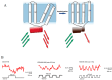
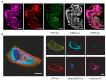
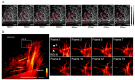
Far-red fluorescent proteins (FPs) have emerged as indispensable tools in in vivo imaging, playing a pivotal role in elucidating fundamental mechanisms and addressing application issues in biotechnology and biomedical fields. Their ability for deep penetration, coupled with reduced light scattering and absorption, robust resistance to autofluorescence, and diminished phototoxicity, has positioned far-red biosensors at the forefront of non-invasive visualization techniques for observing intracellular activities and intercellular behaviors. In this review, far-red FPs and their applications in living systems are mainly discussed. Firstly, various far-red FPs, characterized by emission peaks spanning from 600 nm to 650 nm, are introduced. This is followed by a detailed presentation of the fundamental principles enabling far-red biosensors to detect biomolecules and environmental changes. Furthermore, the review accentuates the superiority of far-red FPs in multi-color imaging. In addition, significant emphasis is placed on the value of far-red FPs in improving imaging resolution, highlighting their great contribution to the advancement of in vivo imaging.
Keywords: far-red fluorescent proteins; fluorescence imaging; fluorescent biosensors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

