Graph-Based Pan-Genome Reveals the Pattern of Deleterious Mutations during the Domestication of Saccharomyces cerevisiae
- PMID: 39194902
- PMCID: PMC11355510
- DOI: 10.3390/jof10080575
Graph-Based Pan-Genome Reveals the Pattern of Deleterious Mutations during the Domestication of Saccharomyces cerevisiae
Abstract
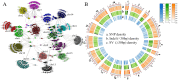
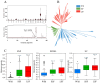
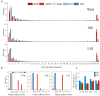
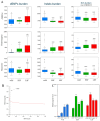
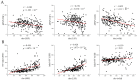
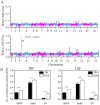
The "cost of domestication" hypothesis suggests that the domestication of wild species increases the number, frequency, and/or proportion of deleterious genetic variants, potentially reducing their fitness in the wild. While extensively studied in domesticated species, this phenomenon remains understudied in fungi. Here, we used Saccharomyces cerevisiae, the world's oldest domesticated fungus, as a model to investigate the genomic characteristics of deleterious variants arising from fungal domestication. Employing a graph-based pan-genome approach, we identified 1,297,761 single nucleotide polymorphisms (SNPs), 278,147 insertion/deletion events (indels; <30 bp), and 19,967 non-redundant structural variants (SVs; ≥30 bp) across 687 S. cerevisiae isolates. Comparing these variants with synonymous SNPs (sSNPs) as neutral controls, we found that the majority of the derived nonsynonymous SNPs (nSNPs), indels, and SVs were deleterious. Heterozygosity was positively correlated with the impact of deleterious SNPs, suggesting a role of genetic diversity in mitigating their effects. The domesticated isolates exhibited a higher additive burden of deleterious SNPs (dSNPs) than the wild isolates, but a lower burden of indels and SVs. Moreover, the domesticated S. cerevisiae showed reduced rates of adaptive evolution relative to the wild S. cerevisiae. In summary, deleterious variants tend to be heterozygous, which may mitigate their harmful effects, but they also constrain breeding potential. Addressing deleterious alleles and minimizing the genetic load are crucial considerations for future S. cerevisiae breeding efforts.
Keywords: Saccharomyces cerevisiae; deleterious mutations; domestication; graph-based pan-genome; heterozygosity.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Larson G., Fuller D.Q. The Evolution of Animal Domestication. Annu. Rev. Ecol. Evol. Syst. 2014;45:115–136. doi: 10.1146/annurev-ecolsys-110512-135813. - DOI
-
- Marsden C.D., Ortega-Del Vecchyo D., O’Brien D.P., Taylor J.F., Ramirez O., Vilà C., Marques-Bonet T., Schnabel R.D., Wayne R.K., Lohmueller K.E. Bottlenecks and Selective Sweeps during Domestication Have Increased Deleterious Genetic Variation in Dogs. Proc. Natl. Acad. Sci. USA. 2016;113:152–157. doi: 10.1073/pnas.1512501113. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

