Functional Diversity and Engineering of the Adenylation Domains in Nonribosomal Peptide Synthetases
- PMID: 39195464
- PMCID: PMC11355689
- DOI: 10.3390/md22080349
Functional Diversity and Engineering of the Adenylation Domains in Nonribosomal Peptide Synthetases
Abstract
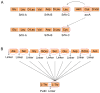
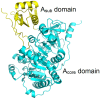
Nonribosomal peptides (NRPs) are biosynthesized by nonribosomal peptide synthetases (NRPSs) and are widely distributed in both terrestrial and marine organisms. Many NRPs and their analogs are biologically active and serve as therapeutic agents. The adenylation (A) domain is a key catalytic domain that primarily controls the sequence of a product during the assembling of NRPs and thus plays a predominant role in the structural diversity of NRPs. Engineering of the A domain to alter substrate specificity is a potential strategy for obtaining novel NRPs for pharmaceutical studies. On the basis of introducing the catalytic mechanism and multiple functions of the A domains, this article systematically describes several representative NRPS engineering strategies targeting the A domain, including mutagenesis of substrate-specificity codes, substitution of condensation-adenylation bidomains, the entire A domain or its subdomains, domain insertion, and whole-module rearrangements.
Keywords: adenylation domain in nonribosomal peptide synthetase; interrupted adenylation domain with methylase activity; nonribosomal peptide synthetase engineering targeting the adenylation domain; substrate-specificity codes of the adenylation domain.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

