A genome-wide CRISPR screen identifies BRD4 as a regulator of cardiomyocyte differentiation
- PMID: 39196112
- PMCID: PMC11361716
- DOI: 10.1038/s44161-024-00431-1
A genome-wide CRISPR screen identifies BRD4 as a regulator of cardiomyocyte differentiation
Abstract
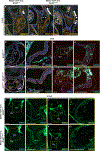
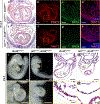
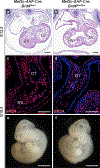
Human induced pluripotent stem cell (hiPSC) to cardiomyocyte (CM) differentiation has reshaped approaches to studying cardiac development and disease. In this study, we employed a genome-wide CRISPR screen in a hiPSC to CM differentiation system and reveal here that BRD4, a member of the bromodomain and extraterminal (BET) family, regulates CM differentiation. Chemical inhibition of BET proteins in mouse embryonic stem cell (mESC)-derived or hiPSC-derived cardiac progenitor cells (CPCs) results in decreased CM differentiation and persistence of cells expressing progenitor markers. In vivo, BRD4 deletion in second heart field (SHF) CPCs results in embryonic or early postnatal lethality, with mutants demonstrating myocardial hypoplasia and an increase in CPCs. Single-cell transcriptomics identified a subpopulation of SHF CPCs that is sensitive to BRD4 loss and associated with attenuated CM lineage-specific gene programs. These results highlight a previously unrecognized role for BRD4 in CM fate determination during development and a heterogenous requirement for BRD4 among SHF CPCs.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests
D.S. is a scientific co-founder, shareholder and director of Tenaya Therapeutics. S.M.H. is an executive, officer and shareholder of Amgen and is a scientific co-founder and shareholder of Tenaya Therapeutics. M.J. is founder, shareholder and executive of Sapient Bioanalytics, LLC. The remaining authors declare no competing interests.
Figures













Comment in
-
Taking BETs on cardiomyocyte differentiation.Nat Cardiovasc Res. 2024 Mar;3(3):265-266. doi: 10.1038/s44161-024-00443-x. Nat Cardiovasc Res. 2024. PMID: 39196116 No abstract available.
References
-
- Van Der Linde D, Konings EEM & Slager M. a., Witsenburg M, Helbing W. a., Takkenberg JJM & Roos-Hesselink JW Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J. Am. Coll. Cardiol. 58, 2241–2247 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
