Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult
- PMID: 39196121
- PMCID: PMC11358021
- DOI: 10.1038/s44161-024-00448-6
Kupffer cells dictate hepatic responses to the atherogenic dyslipidemic insult
Abstract
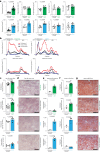
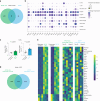
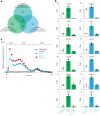
Apolipoprotein-B (APOB)-containing lipoproteins cause atherosclerosis. Whether the vasculature is the initially responding site or if atherogenic dyslipidemia affects other organs simultaneously is unknown. Here we show that the liver responds to a dyslipidemic insult based on inducible models of familial hypercholesterolemia and APOB tracing. An acute transition to atherogenic APOB lipoprotein levels resulted in uptake by Kupffer cells and rapid accumulation of triglycerides and cholesterol in the liver. Bulk and single-cell RNA sequencing revealed a Kupffer-cell-specific transcriptional program that was not activated by a high-fat diet alone or detected in standard liver function or pathological assays, even in the presence of fulminant atherosclerosis. Depletion of Kupffer cells altered the dynamic of plasma and liver lipid concentrations, indicating that these liver macrophages help restrain and buffer atherogenic lipoproteins while simultaneously secreting atherosclerosis-modulating factors into plasma. Our results place Kupffer cells as key sentinels in organizing systemic responses to lipoproteins at the initiation of atherosclerosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures











Comment in
-
Linking MASLD to ACVD through Kupffer cells.Nat Cardiovasc Res. 2024 Mar;3(3):258-259. doi: 10.1038/s44161-024-00442-y. Nat Cardiovasc Res. 2024. PMID: 39196120 No abstract available.
References
-
- Boren, J. et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease: pathophysiological, genetic, and therapeutic insights: a consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J.41, 2313–2330 (2020). 10.1093/eurheartj/ehz962 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
