Semaphorin-3A regulates liver sinusoidal endothelial cell porosity and promotes hepatic steatosis
- PMID: 39196233
- PMCID: PMC11358038
- DOI: 10.1038/s44161-024-00487-z
Semaphorin-3A regulates liver sinusoidal endothelial cell porosity and promotes hepatic steatosis
Abstract
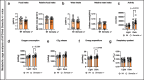
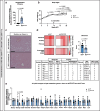
Prevalence of metabolic dysfunction-associated steatotic liver disease (MASLD), formerly known as non-alcoholic fatty liver disease, increases worldwide and associates with type 2 diabetes and other cardiometabolic diseases. Here we demonstrate that Sema3a is elevated in liver sinusoidal endothelial cells of animal models for obesity, type 2 diabetes and MASLD. In primary human liver sinusoidal endothelial cells, saturated fatty acids induce expression of SEMA3A, and loss of a single allele is sufficient to reduce hepatic fat content in diet-induced obese mice. We show that semaphorin-3A regulates the number of fenestrae through a signaling cascade that involves neuropilin-1 and phosphorylation of cofilin-1 by LIM domain kinase 1. Finally, inducible vascular deletion of Sema3a in adult diet-induced obese mice reduces hepatic fat content and elevates very low-density lipoprotein secretion. Thus, we identified a molecular pathway linking hyperlipidemia to microvascular defenestration and early development of MASLD.
© 2024. The Author(s).
Conflict of interest statement
M.R. receives consultation fees from AstraZeneca, Boehringer Ingelheim, Echosens, Madrigal Pharmaceuticals, MSD Sharp & Dohme, Novo Nordisk, Target RWE. The other authors declare no competing interests.
Figures







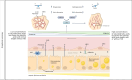


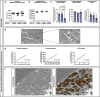

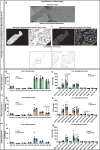
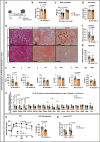
References
Publication types
MeSH terms
Substances
Grants and funding
- La1216/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- RTG 2576 vivid/Deutsche Forschungsgemeinschaft (German Research Foundation)
- GRK2576/Deutsche Forschungsgemeinschaft (German Research Foundation)
- La1216/6-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- IRTG 1902/Deutsche Forschungsgemeinschaft (German Research Foundation)
- Priority Program 2041 (SPP 2041)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- GRK2576/Deutsche Forschungsgemeinschaft (German Research Foundation)
- GRK2576/Deutsche Forschungsgemeinschaft (German Research Foundation)
- GRK2576/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 101147319 (EBRAINS 2.0 Project)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- "CESYDE", Grant agreement No. 759099/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- HORIZON-HLTH-2022-STAYHLTH-02-01: Panel A/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- INTERCEPT-T2D consortium/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
LinkOut - more resources
Full Text Sources
Medical
