Targeted Protein Localization by Covalent 14-3-3 Recruitment
- PMID: 39196545
- PMCID: PMC12011284
- DOI: 10.1021/jacs.3c12389
Targeted Protein Localization by Covalent 14-3-3 Recruitment
Abstract
14-3-3 proteins have a unique ability to bind and sequester a multitude of diverse phosphorylated signaling proteins and transcription factors. Many previous studies have shown that interactions of 14-3-3 with specific phosphorylated substrate proteins can be enhanced through small-molecule natural products or fully synthetic molecular glue interactions. However, enhancing 14-3-3 interactions with both therapeutically intractable transcription factor substrates and potential neo-substrates to sequester and inhibit their function remains elusive. One of the 14-3-3 proteins, 14-3-3σ or SFN, has cysteine C38 at the substrate-binding interface, near the sites where previous 14-3-3 molecular glues have been found to bind. In this study, we screen a fully synthetic cysteine-reactive covalent ligand library to identify molecular glues that enhance the interaction of 14-3-3σ with not only druggable transcription factors such as estrogen receptor (ERα) but also challenging oncogenic transcription factors such as YAP and TAZ, which are part of the Hippo transducer pathway. We identify a hit EN171 that covalently targets both C38 and C96 on 14-3-3 to enhance 14-3-3 interactions with ERα, YAP, and TAZ, leading to impaired estrogen receptor and Hippo pathway transcriptional activity. We further demonstrate that EN171 could not only be used as a molecular glue to enhance native protein interactions but could also be used as a covalent 14-3-3 recruiter in heterobifunctional molecules to sequester nuclear neo-substrates such as BRD4 and BLC6 into the cytosol. Overall, our study reveals a covalent ligand that acts as a novel 14-3-3 molecular glue for challenging transcription factors such as YAP and TAZ and demonstrates that these glues can be potentially utilized in heterobifunctional molecules to sequester nuclear neo-substrates out of the nucleus and into the cytosol to enable targeted protein localization.
Conflict of interest statement
Competing Financial Interests Statement
DKN is a co-founder, shareholder, and scientific advisory board member for Frontier Medicines and Vicinitas Therapeutics. DKN is a member of the board of directors for Vicinitas Therapeutics. DKN is also on the scientific advisory board of The Mark Foundation for Cancer Research, Photys Therapeutics, Oerth Bio, Apertor Pharmaceuticals, and Deciphera Therapeutics. DKN is also an Investment Advisory Partner for a16z Bio, an Advisory Board member for Droia Ventures, and an iPartner for The Column Group.
Figures

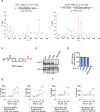

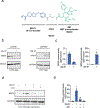
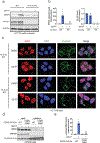

References
-
- Andrei SA et al. Stabilization of protein-protein interactions in drug discovery. Expert Opin. Drug Discov. 12, 925–940 (2017). - PubMed
-
- Milroy L-G, Grossmann TN, Hennig S, Brunsveld L & Ottmann C Modulators of protein-protein interactions. Chem. Rev. 114, 4695–4748 (2014). - PubMed
-
- Ottmann C et al. A structural rationale for selective stabilization of anti-tumor interactions of 14-3-3 proteins by cotylenin A. J. Mol. Biol. 386, 913–919 (2009). - PubMed
-
- Ballone A, Centorrino F, Wolter M & Ottmann C Structural characterization of 14-3-3ζ in complex with the human Son of sevenless homolog 1 (SOS1). J. Struct. Biol. 202, 210–215 (2018). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

