An autoimmune transcriptional circuit drives FOXP3+ regulatory T cell dysfunction
- PMID: 39196959
- PMCID: PMC12051482
- DOI: 10.1126/scitranslmed.adp1720
An autoimmune transcriptional circuit drives FOXP3+ regulatory T cell dysfunction
Abstract
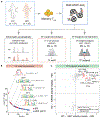
Autoimmune diseases, among the most common disorders of young adults, are mediated by genetic and environmental factors. Although CD4+FOXP3+ regulatory T cells (Tregs) play a central role in preventing autoimmunity, the molecular mechanism underlying their dysfunction is unknown. Here, we performed comprehensive transcriptomic and epigenomic profiling of Tregs in the autoimmune disease multiple sclerosis (MS) to identify critical transcriptional programs regulating human autoimmunity. We found that up-regulation of a primate-specific short isoform of PR domain zinc finger protein 1 (PRDM1-S) induces expression of serum and glucocorticoid-regulated kinase 1 (SGK1) independent from the evolutionarily conserved long PRDM1, which led to destabilization of forkhead box P3 (FOXP3) and Treg dysfunction. This aberrant PRDM1-S/SGK1 axis is shared among other autoimmune diseases. Furthermore, the chromatin landscape profiling in Tregs from individuals with MS revealed enriched activating protein-1 (AP-1)/interferon regulatory factor (IRF) transcription factor binding as candidate upstream regulators of PRDM1-S expression and Treg dysfunction. Our study uncovers a mechanistic model where the evolutionary emergence of PRDM1-S and epigenetic priming of AP-1/IRF may be key drivers of dysfunctional Tregs in autoimmune diseases.
Conflict of interest statement
D.A.H. has received research funding from Bristol-Myers Squibb, Novartis, Sanofi, and Genentech. He has been a consultant for Bayer Pharmaceuticals, Bristol Myers Squibb, Compass Therapeutics, EMD Serono, Genentech, Juno Therapeutics, Novartis Pharmaceuticals, Proclara Biosciences, Sage Therapeutics, and Sanofi Genzyme. B.E.B. declares outside interests in Fulcrum Therapeutics, Arsenal Biosciences, HiFiBio, Cell Signaling Technologies, and Chroma Medicine.
Figures


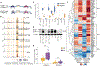


References
-
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA, Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012). - PMC - PubMed
-
- Yoshida H, Lareau CA, Ramirez RN, Rose SA, Maier B, Wroblewska A, Desland F, Chudnovskiy A, Mortha A, Dominguez C, Tellier J, Kim E, Dwyer D, Shinton S, Nabekura T, Qi Y, Yu B, Robinette M, Kim KW, Wagers A, Rhoads A, Nutt SL, Brown BD, Mostafavi S, Buenrostro JD, Benoist C, Immunological Genome Project, The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912.e820 (2019). - PMC - PubMed
-
- Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, Hatan M, Carrasco-Alfonso MJ, Mayer D, Luckey CJ, Patsopoulos NA, De Jager PL, Kuchroo VK, Epstein CB, Daly MJ, Hafler DA, Bernstein BE, Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 (2015). - PMC - PubMed
-
- Hellberg S, Eklund D, Gawel DR, Kopsen M, Zhang H, Nestor CE, Kockum I, Olsson T, Skogh T, Kastbom A, Sjowall C, Vrethem M, Hakansson I, Benson M, Jenmalm MC, Gustafsson M, Ernerudh J, Dynamic response genes in CD4+ T cells reveal a network of interactive proteins that classifies disease activity in multiple sclerosis. Cell Rep. 16, 2928–2939 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

