Single-cell transcriptomic analysis of retinal immune regulation and blood-retinal barrier function during experimental autoimmune uveitis
- PMID: 39198470
- PMCID: PMC11358488
- DOI: 10.1038/s41598-024-68401-y
Single-cell transcriptomic analysis of retinal immune regulation and blood-retinal barrier function during experimental autoimmune uveitis
Abstract
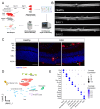
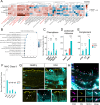
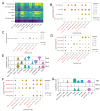
Uveitis is characterised by breakdown of the blood-retinal barrier (BRB), allowing infiltration of immune cells that mediate intraocular inflammation, which can lead to irreversible damage of the neuroretina and the loss of sight. Treatment of uveitis relies heavily on corticosteroids and systemic immunosuppression due to limited understanding of disease pathogenesis. We performed single-cell RNA-sequencing of retinas, as well as bulk RNA-sequencing of retinal pigment epithelial (RPE) cells from mice with experimental autoimmune uveitis (EAU) versus healthy control. This revealed that the Th1/Th17-driven disease induced strong gene expression changes in response to inflammation in rods, cones, Müller glia and RPE. In particular, Müller glia and RPE cells were found to upregulate expression of chemokines, complement factors, leukocyte adhesion molecules and MHC class II, thus highlighting their contributions to immune cell recruitment and antigen presentation at the inner and outer BRB, respectively. Additionally, ligand-receptor interaction analysis with CellPhoneDB revealed key interactions between Müller glia and T cell / natural killer cell subsets via chemokines, galectin-9 to P4HB/TIM-3, PD-L1 to PD-1, and nectin-2/3 to TIGIT signalling axes. Our findings elucidate mechanisms contributing to breakdown of retinal immune privilege during uveitis and identify novel targets for therapeutic interventions.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interests. The views expressed are those of the authors and not necessarily those of the Wellcome Trust, MRC or NIHR.
Figures


References
-
- Fields, M. A., Del Priore, L. V., Adelman, R. A. & Rizzolo, L. J. Interactions of the choroid, Bruch’s membrane, retinal pigment epithelium, and neurosensory retina collaborate to form the outer blood-retinal-barrier. Prog. Retin. Eye Res.76, 100803 (2020). 10.1016/j.preteyeres.2019.100803 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

