Comprehensive serum glycopeptide spectra analysis to identify early-stage epithelial ovarian cancer
- PMID: 39198565
- PMCID: PMC11358426
- DOI: 10.1038/s41598-024-70228-6
Comprehensive serum glycopeptide spectra analysis to identify early-stage epithelial ovarian cancer
Abstract
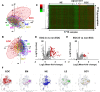
Epithelial ovarian cancer (EOC) is widely recognized as the most lethal gynecological malignancy; however, its early-stage detection remains a considerable clinical challenge. To address this, we have introduced a new method, named Comprehensive Serum Glycopeptide Spectral Analysis (CSGSA), which detects early-stage cancer by combining glycan alterations in serum glycoproteins with tumor markers. We detected 1712 glycopeptides using liquid chromatography-mass spectrometry from the sera obtained from 564 patients with EOC and 1149 controls across 13 institutions. Furthermore, we used a convolutional neural network to analyze the expression patterns of the glycopeptides and tumor markers. Using this approach, we successfully differentiated early-stage EOC (Stage I) from non-EOC, with an area under the curve (AUC) of 0.924 in receiver operating characteristic (ROC) analysis. This method markedly outperforms conventional tumor markers, including cancer antigen 125 (CA125, 0.842) and human epididymis protein 4 (HE4, 0.717). Notably, our method exhibited remarkable efficacy in differentiating early-stage ovarian clear cell carcinoma from endometrioma, achieving a ROC-AUC of 0.808, outperforming CA125 (0.538) and HE4 (0.557). Our study presents a promising breakthrough in the early detection of EOC through the innovative CSGSA method. The integration of glycan alterations with cancer-related tumor markers has demonstrated exceptional diagnostic potential.
Keywords: Clear cell carcinoma; Convolutional neural network; Glycomics; Glycopeptide; Mass spectrometry; Ovarian cancer.
© 2024. The Author(s).
Conflict of interest statement
KT(Kazuhiro Tanabe), TK(Tomoko Katahira), and CH are employed by LSI Medience Corporation, which can provide ovarian cancer screening. MM and LSI Medience Corporation applied for a patent related to this research in Japan (2019–108992). The remaining authors declare that they have no competing interests.
Figures




References
-
- Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. CA: Cancer J. Clin.73, 17–48 (2023). - PubMed
-
- Cancer Research UK, Ovarian cancer survival statistics, https://www.cancerresearchuk.org/about-cancer/ovarian-cancer/survival, accessed on Feb. 14, 2023.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

