Plasmid-encoded insertion sequences promote rapid adaptation in clinical enterobacteria
- PMID: 39198572
- PMCID: PMC7616626
- DOI: 10.1038/s41559-024-02523-4
Plasmid-encoded insertion sequences promote rapid adaptation in clinical enterobacteria
Abstract
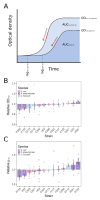
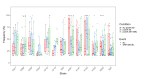
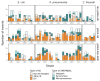
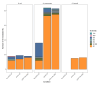
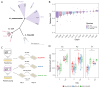
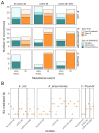
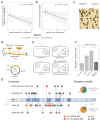
Plasmids are extrachromosomal genetic elements commonly found in bacteria. They are known to fuel bacterial evolution through horizontal gene transfer, and recent analyses indicate that they can also promote intragenomic adaptations. However, the role of plasmids as catalysts of bacterial evolution beyond horizontal gene transfer is poorly explored. In this study, we investigated the impact of a widespread conjugative plasmid, pOXA-48, on the evolution of several multidrug-resistant clinical enterobacteria. Combining experimental and within-patient evolution analyses, we unveiled that plasmid pOXA-48 promotes bacterial evolution through the transposition of plasmid-encoded insertion sequence 1 (IS1) elements. Specifically, IS1-mediated gene inactivation expedites the adaptation rate of clinical strains in vitro and fosters within-patient adaptation in the gut microbiota. We deciphered the mechanism underlying the plasmid-mediated surge in IS1 transposition, revealing a negative feedback loop regulated by the genomic copy number of IS1. Given the overrepresentation of IS elements in bacterial plasmids, our findings suggest that plasmid-mediated IS1 transposition represents a crucial mechanism for swift bacterial adaptation.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures




Update of
-
Plasmid-encoded insertion sequences promote rapid adaptation in clinical enterobacteria.bioRxiv [Preprint]. 2024 Mar 1:2024.03.01.582297. doi: 10.1101/2024.03.01.582297. bioRxiv. 2024. Update in: Nat Ecol Evol. 2024 Nov;8(11):2097-2112. doi: 10.1038/s41559-024-02523-4. PMID: 38903098 Free PMC article. Updated. Preprint.
References
-
- Wiedenbeck J, Cohan FM. Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol Rev. 2011;35:957–976. - PubMed
-
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. - PubMed
-
- Harrison E, Brockhurst MA. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends in Microbiology. 2012;20:262–267. - PubMed
-
- Botelho J, Schulenburg H. The Role of Integrative and Conjugative Elements in Antibiotic Resistance Evolution. Trends in Microbiology. 2021;29:8–18. - PubMed
-
- San Millan A. Evolution of Plasmid-Mediated Antibiotic Resistance in the Clinical Context. Trends in Microbiology. 2018;26:978–985. - PubMed
MeSH terms
Substances
Grants and funding
- PI23/01945/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- CD21/00115/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- 101077809-HorizonGT/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- CP20/00154/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- 895671-REPLAY/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- 757440-PLASREVOLUTION/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- PI19/00749/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- LCF/BQ/PR22/11920001/"la Caixa" Foundation (Caixa Foundation)
- Research Grant 2022/European Society of Clinical Microbiology and Infectious Diseases (ESCMID)
- 757440/ERC_/European Research Council/International
- PI21/01363/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

