Cooperative thalamocortical circuit mechanism for sensory prediction errors
- PMID: 39198646
- PMCID: PMC11390482
- DOI: 10.1038/s41586-024-07851-w
Cooperative thalamocortical circuit mechanism for sensory prediction errors
Abstract
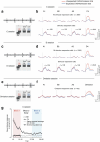
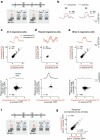
The brain functions as a prediction machine, utilizing an internal model of the world to anticipate sensations and the outcomes of our actions. Discrepancies between expected and actual events, referred to as prediction errors, are leveraged to update the internal model and guide our attention towards unexpected events1-10. Despite the importance of prediction-error signals for various neural computations across the brain, surprisingly little is known about the neural circuit mechanisms responsible for their implementation. Here we describe a thalamocortical disinhibitory circuit that is required for generating sensory prediction-error signals in mouse primary visual cortex (V1). We show that violating animals' predictions by an unexpected visual stimulus preferentially boosts responses of the layer 2/3 V1 neurons that are most selective for that stimulus. Prediction errors specifically amplify the unexpected visual input, rather than representing non-specific surprise or difference signals about how the visual input deviates from the animal's predictions. This selective amplification is implemented by a cooperative mechanism requiring thalamic input from the pulvinar and cortical vasoactive-intestinal-peptide-expressing (VIP) inhibitory interneurons. In response to prediction errors, VIP neurons inhibit a specific subpopulation of somatostatin-expressing inhibitory interneurons that gate excitatory pulvinar input to V1, resulting in specific pulvinar-driven response amplification of the most stimulus-selective neurons in V1. Therefore, the brain prioritizes unpredicted sensory information by selectively increasing the salience of unpredicted sensory features through the synergistic interaction of thalamic input and neocortical disinhibitory circuits.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures















References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

