Prokaryotic-virus-encoded auxiliary metabolic genes throughout the global oceans
- PMID: 39198891
- PMCID: PMC11360552
- DOI: 10.1186/s40168-024-01876-z
Prokaryotic-virus-encoded auxiliary metabolic genes throughout the global oceans
Abstract
Background: Prokaryotic microbes have impacted marine biogeochemical cycles for billions of years. Viruses also impact these cycles, through lysis, horizontal gene transfer, and encoding and expressing genes that contribute to metabolic reprogramming of prokaryotic cells. While this impact is difficult to quantify in nature, we hypothesized that it can be examined by surveying virus-encoded auxiliary metabolic genes (AMGs) and assessing their ecological context.
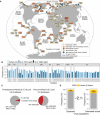
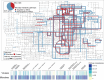
Results: We systematically developed a global ocean AMG catalog by integrating previously described and newly identified AMGs and then placed this catalog into ecological and metabolic contexts relevant to ocean biogeochemistry. From 7.6 terabases of Tara Oceans paired prokaryote- and virus-enriched metagenomic sequence data, we increased known ocean virus populations to 579,904 (up 16%). From these virus populations, we then conservatively identified 86,913 AMGs that grouped into 22,779 sequence-based gene clusters, 7248 (~ 32%) of which were not previously reported. Using our catalog and modeled data from mock communities, we estimate that ~ 19% of ocean virus populations carry at least one AMG. To understand AMGs in their metabolic context, we identified 340 metabolic pathways encoded by ocean microbes and showed that AMGs map to 128 of them. Furthermore, we identified metabolic "hot spots" targeted by virus AMGs, including nine pathways where most steps (≥ 0.75) were AMG-targeted (involved in carbohydrate, amino acid, fatty acid, and nucleotide metabolism), as well as other pathways where virus-encoded AMGs outnumbered cellular homologs (involved in lipid A phosphates, phosphatidylethanolamine, creatine biosynthesis, phosphoribosylamine-glycine ligase, and carbamoyl-phosphate synthase pathways).
Conclusions: Together, this systematically curated, global ocean AMG catalog and analyses provide a valuable resource and foundational observations to understand the role of viruses in modulating global ocean metabolisms and their biogeochemical implications. Video Abstract.
Keywords: Tara Oceans; AMGs; Prokaryotic.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

