Integrated Multi-Omics Analysis of Cerebrospinal Fluid in Postoperative Delirium
- PMID: 39199312
- PMCID: PMC11352186
- DOI: 10.3390/biom14080924
Integrated Multi-Omics Analysis of Cerebrospinal Fluid in Postoperative Delirium
Abstract
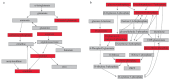
Preoperative risk biomarkers for delirium may aid in identifying high-risk patients and developing intervention therapies, which would minimize the health and economic burden of postoperative delirium. Previous studies have typically used single omics approaches to identify such biomarkers. Preoperative cerebrospinal fluid (CSF) from the Healthier Postoperative Recovery study of adults ≥ 63 years old undergoing elective major orthopedic surgery was used in a matched pair delirium case-no delirium control design. We performed metabolomics and lipidomics, which were combined with our previously reported proteomics results on the same samples. Differential expression, clustering, classification, and systems biology analyses were applied to individual and combined omics datasets. Probabilistic graph models were used to identify an integrated multi-omics interaction network, which included clusters of heterogeneous omics interactions among lipids, metabolites, and proteins. The combined multi-omics signature of 25 molecules attained an AUC of 0.96 [95% CI: 0.85-1.00], showing improvement over individual omics-based classification. We conclude that multi-omics integration of preoperative CSF identifies potential risk markers for delirium and generates new insights into the complex pathways associated with delirium. With future validation, this hypotheses-generating study may serve to build robust biomarkers for delirium and improve our understanding of its pathophysiology.
Keywords: delirium; lipidomics; metabolomics; multi-omics; proteomics; risk factors.
Conflict of interest statement
The funders had no role in the design or conduct of the study; collection, management, analysis, or interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication. The authors report no biomedical financial interests or potential conflicts of interest. Zhongcong Xie provided consulting service to Baxter, NanoMosaic, Shanghai 4th, 9th and 10th hospitals, Shanghai Mental Health Center of Shanghai Jiao Tong University School of Medicine, and <<Anesthesiology and Perioperative Science>> in last 36 months.
Figures





References
-
- Gou R.Y., Hshieh T.T., Marcantonio E.R., Cooper Z., Jones R.N., Travison T.G., Fong T.G., Abdeen A., Lange J., Earp B., et al. One-year Medicare costs associated with delirium in older patients undergoing major elective surgery. JAMA Surg. 2021;156:430–442. doi: 10.1001/jamasurg.2020.7260. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 AG051658/AG/NIA NIH HHS/United States
- NA/Beth Israel Deaconess Medical Center
- R21 AG081763/AG/NIA NIH HHS/United States
- R03 AG061582/AG/NIA NIH HHS/United States
- K24 AG035075/AG/NIA NIH HHS/United States
- 5P01CA120964 and 5P30CA006516/NH/NIH HHS/United States
- K01 AG057836/AG/NIA NIH HHS/United States
- R24 AG054259/AG/NIA NIH HHS/United States
- P01 AG031720/AG/NIA NIH HHS/United States
- R01AG051658, P01AG031720, R33AG071744, K01AG057836, K24AG035075, R03AG061582, R21AG081763/AG/NIA NIH HHS/United States
- R01 AG079864/AG/NIA NIH HHS/United States
- R33 AG071744/AG/NIA NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- AARF-18-560786/ALZ/Alzheimer's Association/United States
- P01 CA120964/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

