RNA Surveillance Factor SMG5 Is Essential for Mouse Embryonic Stem Cell Differentiation
- PMID: 39199410
- PMCID: PMC11352633
- DOI: 10.3390/biom14081023
RNA Surveillance Factor SMG5 Is Essential for Mouse Embryonic Stem Cell Differentiation
Abstract
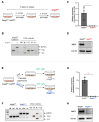
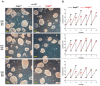
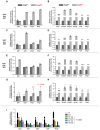
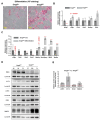
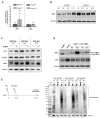
Nonsense-mediated mRNA decay (NMD) is a highly conserved post-transcriptional gene expression regulatory mechanism in eukaryotic cells. NMD eliminates aberrant mRNAs with premature termination codons to surveil transcriptome integrity. Furthermore, NMD fine-tunes gene expression by destabilizing RNAs with specific NMD features. Thus, by controlling the quality and quantity of the transcriptome, NMD plays a vital role in mammalian development, stress response, and tumorigenesis. Deficiencies of NMD factors result in early embryonic lethality, while the underlying mechanisms are poorly understood. SMG5 is a key NMD factor. In this study, we generated an Smg5 conditional knockout mouse model and found that Smg5-null results in early embryonic lethality before E13.5. Furthermore, we produced multiple lines of Smg5 knockout mouse embryonic stem cells (mESCs) and found that the deletion of Smg5 in mESCs does not compromise cell viability. Smg5-null delays differentiation of mESCs. Mechanistically, our study reveals that the c-MYC protein, but not c-Myc mRNA, is upregulated in SMG5-deficient mESCs. The overproduction of c-MYC protein could be caused by enhanced protein synthesis upon SMG5 loss. Furthermore, SMG5-null results in dysregulation of alternative splicing on multiple stem cell differentiation regulators. Overall, our findings underscore the importance of SMG5-NMD in regulating mESC cell-state transition.
Keywords: SMG5; c-MYC; differentiation; mouse embryonic stem cells; nonsense-mediated mRNA decay.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
SMG5, a component of nonsense-mediated mRNA decay, is essential for the mouse spermatogonial differentiation and maintenance.FASEB J. 2024 Dec 13;38(24):e70268. doi: 10.1096/fj.202402422R. FASEB J. 2024. PMID: 39704269
-
Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells.RNA. 2018 Apr;24(4):557-573. doi: 10.1261/rna.063719.117. Epub 2018 Jan 18. RNA. 2018. PMID: 29348139 Free PMC article.
-
MicroRNA 433 regulates nonsense-mediated mRNA decay by targeting SMG5 mRNA.BMC Mol Biol. 2016 Jul 29;17(1):17. doi: 10.1186/s12867-016-0070-z. BMC Mol Biol. 2016. PMID: 27473591 Free PMC article.
-
Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression.Semin Cell Dev Biol. 2018 Mar;75:78-87. doi: 10.1016/j.semcdb.2017.08.053. Epub 2017 Sep 1. Semin Cell Dev Biol. 2018. PMID: 28866327 Review.
-
[Advances in the roles and mechanisms of nonsense-mediated mRNA decay in embryonic development].Sheng Li Xue Bao. 2019 Apr 25;71(2):327-335. Sheng Li Xue Bao. 2019. PMID: 31008493 Review. Chinese.
Cited by
-
Fine-tuning of Wnt signaling by RNA surveillance factor Smg5 in the mouse craniofacial development.iScience. 2025 Feb 6;28(3):111972. doi: 10.1016/j.isci.2025.111972. eCollection 2025 Mar 21. iScience. 2025. PMID: 40071146 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

