The Personalized Inherited Signature Predisposing to Non-Small-Cell Lung Cancer in Non-Smokers
- PMID: 39199663
- PMCID: PMC11352340
- DOI: 10.3390/cancers16162887
The Personalized Inherited Signature Predisposing to Non-Small-Cell Lung Cancer in Non-Smokers
Abstract
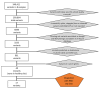
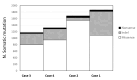
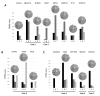
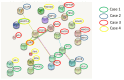
Lung cancer (LC) continues to be an important public health problem, being the most common form of cancer and a major cause of cancer deaths worldwide. Despite the great bulk of research to identify genetic susceptibility genes by genome-wide association studies, only few loci associated to nicotine dependence have been consistently replicated. Our previously published study in few phenotypically discordant sib-pairs identified a combination of germline truncating mutations in known cancer susceptibility genes in never-smoker early-onset LC patients, which does not present in their healthy sib. These results firstly demonstrated the presence of an oligogenic combination of disrupted cancer-predisposing genes in non-smokers patients, giving experimental support to a model of a "private genetic epidemiology". Here, we used a combination of whole-exome and RNA sequencing coupled with a discordant sib's model in a novel cohort of pairs of never-smokers early-onset LC patients and in their healthy sibs used as controls. We selected rare germline variants predicted as deleterious by CADD and SVM bioinformatics tools and absent in the healthy sib. Overall, we identified an average of 200 variants per patient, about 10 of which in cancer-predisposing genes. In most of them, RNA sequencing data reinforced the pathogenic role of the identified variants showing: (i) downregulation in LC tissue (indicating a "second hit" in tumor suppressor genes); (ii) upregulation in cancer tissue (likely oncogene); and (iii) downregulation in both normal and cancer tissue (indicating transcript instability). The combination of the two techniques demonstrates that each patient has an average of six (with a range from four to eight) private mutations with a functional effect in tumor-predisposing genes. The presence of a unique combination of disrupting events in the affected subjects may explain the absence of the familial clustering of non-small-cell lung cancer. In conclusion, these findings indicate that each patient has his/her own "predisposing signature" to cancer development and suggest the use of personalized therapeutic strategies in lung cancer.
Keywords: germline variants; lung cancer susceptibility; next-generation sequencing; oligogenic model; whole-exome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Wild C., Weiderpass E., Stewart B.W. World Cancer Report: Cancer Research for Cancer Prevention 2020. International Agency for Research on Cancer; Lyon, France: 2020.
Grants and funding
LinkOut - more resources
Full Text Sources

