First Data on WGS-Based Typing and Antimicrobial Resistance of Human Salmonella Enteritidis Isolates in Greece
- PMID: 39200008
- PMCID: PMC11350896
- DOI: 10.3390/antibiotics13080708
First Data on WGS-Based Typing and Antimicrobial Resistance of Human Salmonella Enteritidis Isolates in Greece
Abstract
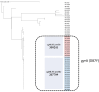
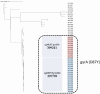
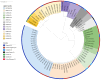
Salmonella enterica subsp. enterica serotype Enteritidis (S. Enteritidis) is one of the major causes of foodborne infections and is responsible for many national and multi-country foodborne outbreaks worldwide. In Greece, human salmonellosis is a mandatory notifiable disease, with laboratory surveillance being on a voluntary basis. This study aims to provide the first insights into the genetic characteristics and antimicrobial resistance profiles of 47 S. Enteritidis human isolates using whole-genome sequencing (WGS) technology. The S. Enteritidis population was mainly resistant to fluoroquinolones due to gyrA point mutations, whereas one isolate presented a multi-resistant plasmid-mediated phenotype. ST11 was the most frequent sequence type, and phylogenetic analysis through the cgMLST and SNP methods revealed considerable genetic diversity. Regarding virulence factors, 8 out of the 24 known SPIs and C63PI were detected. Due to the observed variability between countries, it is of utmost importance to record the circulating S. Enteritidis strains' structure and genomic epidemiology at the national level. WGS is a valuable tool that is revolutionizing our approach to Salmonella by providing a deeper understanding of these pathogens and their impact on human health.
Keywords: S. Enteritidis; phylogenetic analysis; surveillance; whole-genome sequence-based typing.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- WHO . Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007–2015. World Health Organization; Geneva, Switzerland: 2015.
-
- Kadhum W.R., Sviridova L., Snegirev D. The analysis of Salmonella’s ability to survive in different external environments. Biostat. Epidemiol. 2023;7:e2265277. doi: 10.1080/24709360.2023.2265277. - DOI
-
- European Centre for Disease Prevention and Control Salmonellosis. [(accessed on 10 June 2024)]. Available online: https://www.ecdc.europa.eu/en/salmonellosis.
LinkOut - more resources
Full Text Sources

