Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis
- PMID: 39200025
- PMCID: PMC11350675
- DOI: 10.3390/antibiotics13080725
Identifying Cell-Penetrating Peptides for Effectively Delivering Antimicrobial Molecules into Streptococcus suis
Abstract
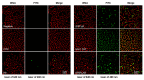
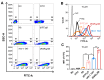
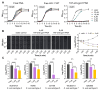
Cell-penetrating peptides (CPPs) are promising carriers to effectively transport antisense oligonucleotides (ASOs), including peptide nucleic acids (PNAs), into bacterial cells to combat multidrug-resistant bacterial infections, demonstrating significant therapeutic potential. Streptococcus suis, a Gram-positive bacterium, is a major bacterial pathogen in pigs and an emerging zoonotic pathogen. In this study, through the combination of super-resolution structured illumination microscopy (SR-SIM), flow cytometry analysis, and toxicity analysis assays, we investigated the suitability of four CPPs for delivering PNAs into S. suis cells: HIV-1 TAT efficiently penetrated S. suis cells with low toxicity against S. suis; (RXR)4XB had high penetration efficiency with inherent toxicity against S. suis; (KFF)3K showed lower penetration efficiency than HIV-1 TAT and (RXR)4XB; K8 failed to penetrate S. suis cells. HIV-1 TAT-conjugated PNA specific for the essential gyrase A subunit gene (TAT-anti-gyrA PNA) effectively inhibited the growth of S. suis. TAT-anti-gyrA PNA exhibited a significant bactericidal effect on serotypes 2, 4, 5, 7, and 9 strains of S. suis, which are known to cause human infections. Our study demonstrates the potential of CPP-ASO conjugates as new antimicrobial compounds for combating S. suis infections. Furthermore, our findings demonstrate that applying SR-SIM and flow cytometry analysis provides a convenient, intuitive, and cost-effective approach to identifying suitable CPPs for delivering cargo molecules into bacterial cells.
Keywords: Streptococcus suis; antisense oligonucleotides; cell-penetrating peptides; peptide nucleic acids.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Antimicrobial Activity of Peptide-Coupled Antisense Peptide Nucleic Acids in Streptococcus pneumoniae.Microbiol Spectr. 2022 Dec 21;10(6):e0049722. doi: 10.1128/spectrum.00497-22. Epub 2022 Nov 2. Microbiol Spectr. 2022. PMID: 36321914 Free PMC article.
-
Influence of Different Cell-Penetrating Peptides on the Antimicrobial Efficiency of PNAs in Streptococcus pyogenes.Mol Ther Nucleic Acids. 2019 Dec 6;18:444-454. doi: 10.1016/j.omtn.2019.09.010. Epub 2019 Sep 19. Mol Ther Nucleic Acids. 2019. PMID: 31655262 Free PMC article.
-
Pyrenebutyrate Enhances the Antibacterial Effect of Peptide-Coupled Antisense Peptide Nucleic Acids in Streptococcus pyogenes.Microorganisms. 2023 Aug 22;11(9):2131. doi: 10.3390/microorganisms11092131. Microorganisms. 2023. PMID: 37763975 Free PMC article.
-
Developments in Cell-Penetrating Peptides as Antiviral Agents and as Vehicles for Delivery of Peptide Nucleic Acid Targeting Hepadnaviral Replication Pathway.Biomolecules. 2018 Jul 16;8(3):55. doi: 10.3390/biom8030055. Biomolecules. 2018. PMID: 30013006 Free PMC article. Review.
-
Streptococcus suis: a re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II - Pathogenesis.Ann Agric Environ Med. 2018 Mar 14;25(1):186-203. doi: 10.26444/aaem/85651. Epub 2018 Mar 2. Ann Agric Environ Med. 2018. PMID: 29575852 Review.
Cited by
-
Probing Peptide Assembly and Interaction via High-Resolution Imaging Techniques: A Mini Review.Int J Mol Sci. 2025 Apr 23;26(9):3998. doi: 10.3390/ijms26093998. Int J Mol Sci. 2025. PMID: 40362238 Free PMC article. Review.
-
Identification of SepF in Streptococcus suis involving cell division.BMC Microbiol. 2025 Mar 31;25(1):179. doi: 10.1186/s12866-025-03919-3. BMC Microbiol. 2025. PMID: 40165076 Free PMC article.
-
Combinatorial Effects of CPP-Modified Antimicrobial Peptides: Synergistic and Additive Interactions Against Pathogenic Bacteria.Int J Mol Sci. 2025 Jun 21;26(13):5968. doi: 10.3390/ijms26135968. Int J Mol Sci. 2025. PMID: 40649746 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

