Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria
- PMID: 39200050
- PMCID: PMC11350732
- DOI: 10.3390/antibiotics13080750
Regulation of Antibiotic Resistance Genes on Agricultural Land Is Dependent on Both Choice of Organic Amendment and Prevalence of Predatory Bacteria
Abstract
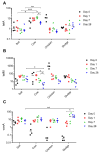
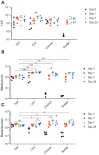
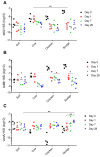
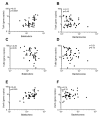
Antibiotic resistance genes (ARGs) are widespread in the environment, and soils, specifically, are hotspots for microorganisms with inherent antibiotic resistance. Manure and sludge used as fertilizers in agricultural production have been shown to contain vast amounts of ARGs, and due to continued applications, ARGs accumulate in agricultural soils. Some soils, however, harbor a resilience capacity that could depend on specific soil properties, as well as the presence of predatory bacteria that are able to hydrolyse living bacteria, including bacteria of clinical importance. The objectives of this study were to (i) investigate if the antibiotic resistance profile of the soil microbiota could be differently affected by the addition of cow manure, chicken manure, and sludge, and (ii) investigate if the amendments had an effect on the presence of predatory bacteria. The three organic amendments were mixed separately with a field soil, divided into pots, and incubated in a greenhouse for 28 days. Droplet digital PCR (ddPCR) was used to quantify three ARGs, two predatory bacteria, and total number of bacteria. In this study, we demonstrated that the choice of organic amendment significantly affected the antibiotic resistance profile of soil, and promoted the growth of predatory bacteria, while the total number of bacteria was unaffected.
Keywords: BALO; antibiotic resistance; manure; sewage sludge; soil; tetracycline; vancomycin.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Antibiotic Resistance in Agricultural Soil and Crops Associated to the Application of Cow Manure-Derived Amendments From Conventional and Organic Livestock Farms.Front Vet Sci. 2021 Feb 23;8:633858. doi: 10.3389/fvets.2021.633858. eCollection 2021. Front Vet Sci. 2021. PMID: 33708812 Free PMC article.
-
Fate of tetracycline and sulfonamide resistance genes in a grassland soil amended with different organic fertilizers.Ecotoxicol Environ Saf. 2019 Apr 15;170:39-46. doi: 10.1016/j.ecoenv.2018.11.059. Epub 2018 Dec 1. Ecotoxicol Environ Saf. 2019. PMID: 30513413
-
Manure-Based Amendments Influence Surface-Associated Bacteria and Markers of Antibiotic Resistance on Radishes Grown in Soils with Different Textures.Appl Environ Microbiol. 2021 Apr 27;87(10):e02753-20. doi: 10.1128/AEM.02753-20. Print 2021 Apr 27. Appl Environ Microbiol. 2021. PMID: 33712421 Free PMC article.
-
Effects of Dairy Manure-Based Amendments and Soil Texture on Lettuce- and Radish-Associated Microbiota and Resistomes.mSphere. 2019 May 8;4(3):e00239-19. doi: 10.1128/mSphere.00239-19. mSphere. 2019. PMID: 31068435 Free PMC article.
-
Field-based evidence for the enrichment of intrinsic antibiotic resistome stimulated by plant-derived fertilizer in agricultural soil.J Environ Sci (China). 2024 Jan;135:728-740. doi: 10.1016/j.jes.2022.08.009. Epub 2022 Aug 14. J Environ Sci (China). 2024. PMID: 37778843
References
-
- He Y., Yuan Q., Mathieu J., Stadler L., Senehi N., Sun R., Alvarez P.J.J. Antibiotic Resistance Genes from Livestock Waste: Occurrence, Dissemination, and Treatment. npj Clean Water. 2020;3:4. doi: 10.1038/s41545-020-0051-0. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

