Phylogenetic Diversity, Antibiotic Resistance, and Virulence of Escherichia coli Strains from Urinary Tract Infections in Algeria
- PMID: 39200073
- PMCID: PMC11350822
- DOI: 10.3390/antibiotics13080773
Phylogenetic Diversity, Antibiotic Resistance, and Virulence of Escherichia coli Strains from Urinary Tract Infections in Algeria
Abstract
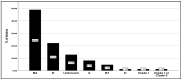
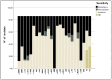
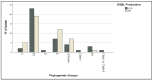
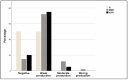
Urinary tract infections (UTIs) caused by Escherichia coli represent a significant public health concern due to the high virulence and antimicrobial resistance exhibited by these pathogens. This study aimed to analyze the phylogenetic diversity and antibiotic resistance profiles of Uropathogenic E. coli (UPEC) strains isolated from UTI patients in Algeria, focusing on virulence factors such as extended β-lactamase (ESBL) production, biofilm formation, and hemolytic activity. Phylogenetic grouping of 86 clinical imipenem resistant E. coli isolates showed the prevalence of group B2 (48.9%), followed by groups E (22.1%), unknown (12.8%), A (8.1%), and B1 (4.7%), and Clade I, D, Clade I, or Clade II (1.2%). The highest resistance rates were observed towards amoxicillin (86.04%), ticarcillin (82.55%), piperacillin (73.25%), nitrofurantoin (84.88%), and trimethoprim-sulfamethoxazole (51.16%). Notably, 69.8% of UPEC strains were multidrug-resistant (MDR) and 23.2% were extensively drug-resistant (XDR). Additionally, 48.9%, 42%, and 71% of strains demonstrated ESBL production, hemolytic activity, and weak biofilm production, respectively. Continuous monitoring and characterization of UPEC strains are essential to track the spread of the most resistant and virulent phylogenetic groups over time, facilitating rapid therapeutic decisions to treat infections and prevent the emergence of new resistant organisms, helping choose the most effective antibiotics and reducing treatment failure.
Keywords: Escherichia coli; antibiotic resistance; biofilm; hemolysin; imipenem; phylogenetic groups; urinary tract infections.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Coura F.M., Diniz A.N., Oliveira Junior C.A., Lage A.P., Lobato F.C.F., Heinemann M.B., Silva R.O.S. Detection of Virulence Genes and the Phylogenetic Groups of Escherichia coli Isolated from Dogs in Brazil. Ciênc. Rural. 2018;48:e20170478. doi: 10.1590/0103-8478cr20170478. - DOI
-
- Bozorgomid A., Chegene Lorestani R., Rostamian M., Nemati Zargaran F., Shahvaisi-Zadeh Z., Akya A. Antibiotic Resistance, Virulence Factors, and Phylogenetic Groups of Escherichia coli Isolated from Hospital Wastewater: A Case Study in the West of Iran. Environ. Health Eng. Manag. 2023;10:131–139. doi: 10.34172/EHEM.2023.15. - DOI
-
- Boroumand M., Naghmachi M., Ghatee M.A. Detection of Phylogenetic Groups and Drug Resistance Genes of Escherichia coli Causing Urinary Tract Infection in Southwest Iran. Jundishapur J. Microbiol. 2021;14:e112547. doi: 10.5812/jjm.112547. - DOI
Grants and funding
- CN00000033/European Commission NextGenerationEU, Piano Nazionale Resistenza e Resilienza (PNRR) - Missione 4 Componente 2 Investimento 1.4 - Avviso N. 3138 del 16 dicembre 2021 rettificato con D.D. n.3175 del 18 dicembre 2021 del Ministero dell'Università e della Ri
- n. IR0000005PO/European Commission - NextGenerationEU, Project SUS-MIRRI.IT "Strengthening the MIRRI Italian Research Infrastructure for Sustainable Bioscience and Bioeconomy"
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

