Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes
- PMID: 39200230
- PMCID: PMC11351110
- DOI: 10.3390/biomedicines12081766
Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes
Abstract
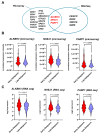
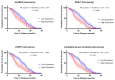
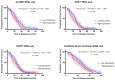
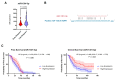
Genomic instability is one of the main drivers of tumorigenesis and the development of hematological malignancies. Cancer cells can remedy chemotherapeutic-induced DNA damage by upregulating DNA-repair genes and ultimately inducing therapy resistance. Nevertheless, the association between the DNA-repair genes, drug resistance, and disease relapse has not been well characterized in acute lymphoblastic leukemia (ALL). This study aimed to explore the role of the DNA-repair machinery and the molecular mechanisms by which it is regulated in early- and late-relapsing pediatric ALL patients. We performed secondary data analysis on the Therapeutically Applicable Research to Generate Effective Treatments (TARGET)-ALL expansion phase II trial of 198 relapsed pediatric precursor B-cell ALL. Comprehensive genetic and epigenetic investigations of 147 DNA-repair genes were conducted in the study. Gene expression was assessed using Microarray and RNA-sequencing platforms. Genomic alternations, methylation status, and miRNA transcriptome were investigated for the candidate DNA-repair genes. We identified three DNA-repair genes, ALKBH3, NHEJ1, and PARP1, that were upregulated in early relapsers compared to late relapsers (p < 0.05). Such upregulation at diagnosis was significantly associated with disease-free survival and overall survival in precursor-B-ALL (p < 0.05). Moreover, PARP1 upregulation accompanied a significant downregulation of its targeting miRNA, miR-1301-3p (p = 0.0152), which was strongly linked with poorer disease-free and overall survivals. Upregulation of DNA-repair genes, PARP1 in particular, increases the likelihood of early relapse of precursor-B-ALL in children. The observation that PARP1 was upregulated in early relapsers relative to late relapsers might serve as a valid rationale for proposing alternative treatment approaches, such as using PARP inhibitors with chemotherapy.
Keywords: DNA repair; PARP1; acute lymphoblastic leukemia; childhood ALL; early relapse; late relapse; miRNA; precursor-B-ALL; prognostic biomarker.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Nabizadeh F., Momtaz S., Ghanbari-Movahed M., Qalekhani F., Mohsenpour H., Aneva I.Y., Bishayee A., Farzaei M.H., Bishayee A. Pediatric Acute Lymphoblastic Leukemia Management Using Multitargeting Bioactive Natural Compounds: A Systematic and Critical Review. Pharmacol. Res. 2022;177:106116. doi: 10.1016/j.phrs.2022.106116. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

