Application of PPAR Ligands and Nanoparticle Technology in Metabolic Steatohepatitis Treatment
- PMID: 39200340
- PMCID: PMC11351628
- DOI: 10.3390/biomedicines12081876
Application of PPAR Ligands and Nanoparticle Technology in Metabolic Steatohepatitis Treatment
Abstract
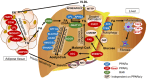
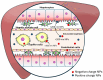
Metabolic dysfunction-associated steatotic liver disease/steatohepatitis (MASLD/MASH) is a major disease worldwide whose effective treatment is challenging. Peroxisome proliferator-activated receptors (PPARs) belong to the nuclear receptor superfamily and function as ligand-activated transcription factors. To date, three distinct subtypes of PPARs have been characterized: PPARα, PPARβ/δ, and PPARγ. PPARα and PPARγ are crucial regulators of lipid metabolism that modulate the transcription of genes involved in fatty acid (FA), bile acid, and cholesterol metabolism. Many PPAR agonists, including natural (FAs, eicosanoids, and phospholipids) and synthetic (fibrate, thiazolidinedione, glitazar, and elafibranor) agonists, have been developed. Furthermore, recent advancements in nanoparticles (NPs) have led to the development of new strategies for MASLD/MASH therapy. This review discusses the applications of specific cell-targeted NPs and highlights the potential of PPARα- and PPARγ-targeted NP drug delivery systems for MASLD/MASH treatment.
Keywords: MASH; MASLD; PPAR; drug delivery; nanoparticles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Peroxisome proliferator-activated receptor (PPAR) alpha and PPARbeta/delta, but not PPARgamma, modulate the expression of genes involved in cardiac lipid metabolism.Circ Res. 2003 Mar 21;92(5):518-24. doi: 10.1161/01.RES.0000060700.55247.7C. Epub 2003 Feb 6. Circ Res. 2003. PMID: 12600885
-
Impact of Peroxisome Proliferator-Activated Receptor Agonists on Myosteatosis in the Context of Metabolic Dysfunction-Associated Steatotic Liver Disease.Discov Med. 2024 Jun;36(185):1139-1153. doi: 10.24976/Discov.Med.202436185.104. Discov Med. 2024. PMID: 38926100 Review.
-
Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells.Circ Res. 2004 May 14;94(9):1168-78. doi: 10.1161/01.RES.0000127122.22685.0A. Circ Res. 2004. PMID: 15142970 Review.
-
Peroxisome proliferator-activated receptors in vascular biology-molecular mechanisms and clinical implications.Vascul Pharmacol. 2006 Jul;45(1):19-28. doi: 10.1016/j.vph.2005.11.014. Epub 2006 Jun 16. Vascul Pharmacol. 2006. PMID: 16782410 Review.
-
The role of peroxisome proliferator-activated receptors in healthy and diseased eyes.Exp Eye Res. 2021 Jul;208:108617. doi: 10.1016/j.exer.2021.108617. Epub 2021 May 16. Exp Eye Res. 2021. PMID: 34010603 Free PMC article. Review.
References
-
- Estes C., Anstee Q.M., Arias-Loste M.T., Bantel H., Bellentani S., Caballeria J., Colombo M., Craxi A., Crespo J., Day C.P., et al. Modeling NAFLD disease burden in China, France, Germany, Italy, Japan, Spain, United Kingdom, and United States for the period 2016–2030. J. Hepatol. 2018;69:896–904. doi: 10.1016/j.jhep.2018.05.036. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

