The Microbiome in Inflammatory Bowel Disease
- PMID: 39200765
- PMCID: PMC11354561
- DOI: 10.3390/jcm13164622
The Microbiome in Inflammatory Bowel Disease
Abstract
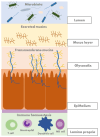
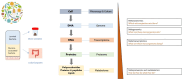
The management of patients with inflammatory bowel disease (IBD) aims to control inflammation through the use of immunosuppressive treatments that target various points in the inflammatory cascade. However, the efficacy of these therapies in the long term is limited, and they often are associated with severe side effects. Although the pathophysiology of the disease is not completely understood, IBD is regarded as a multifactorial disease that occurs due to an inappropriate immune response in genetically susceptible individuals. The gut microbiome is considered one of the main actors in the development of IBD. Gut dysbiosis, characterised by significant changes in the composition and functionality of the gut microbiota, often leads to a reduction in bacterial diversity and anti-inflammatory anaerobic bacteria. At the same time, bacteria with pro-inflammatory potential increase. Although changes in microbiome composition upon biological agent usage have been observed, their role as biomarkers is still unclear. While most studies on IBD focus on the intestinal bacterial population, recent studies have highlighted the importance of other microbial populations, such as viruses and fungi, in gut dysbiosis. In order to modulate the aberrant immune response in patients with IBD, researchers have developed therapies that target different players in the gut microbiome. These innovative approaches hold promise for the future of IBD treatment, although safety concerns are the main limitations, as their effects on humans remain unknown.
Keywords: Crohn’s disease; biofilm; biomarker; dysbiosis; microbiota; ulcerative colitis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources

