Trajectory Analysis in Single-Particle Tracking: From Mean Squared Displacement to Machine Learning Approaches
- PMID: 39201346
- PMCID: PMC11354962
- DOI: 10.3390/ijms25168660
Trajectory Analysis in Single-Particle Tracking: From Mean Squared Displacement to Machine Learning Approaches
Abstract
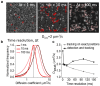
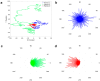
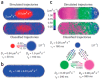
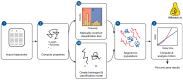
Single-particle tracking is a powerful technique to investigate the motion of molecules or particles. Here, we review the methods for analyzing the reconstructed trajectories, a fundamental step for deciphering the underlying mechanisms driving the motion. First, we review the traditional analysis based on the mean squared displacement (MSD), highlighting the sometimes-neglected factors potentially affecting the accuracy of the results. We then report methods that exploit the distribution of parameters other than displacements, e.g., angles, velocities, and times and probabilities of reaching a target, discussing how they are more sensitive in characterizing heterogeneities and transient behaviors masked in the MSD analysis. Hidden Markov Models are also used for this purpose, and these allow for the identification of different states, their populations and the switching kinetics. Finally, we discuss a rapidly expanding field-trajectory analysis based on machine learning. Various approaches, from random forest to deep learning, are used to classify trajectory motions, which can be identified by motion models or by model-free sets of trajectory features, either previously defined or automatically identified by the algorithms. We also review free software available for some of the analysis methods. We emphasize that approaches based on a combination of the different methods, including classical statistics and machine learning, may be the way to obtain the most informative and accurate results.
Keywords: hidden Markov models; machine learning in biology; molecular diffusion; molecular trajectory statistics; moment scaling spectrum; particle dynamics; quantitative biology; quantitative microscopy; single molecule tracking; single-molecule analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Classification-based motion analysis of single-molecule trajectories using DiffusionLab.Sci Rep. 2022 Jun 10;12(1):9595. doi: 10.1038/s41598-022-13446-0. Sci Rep. 2022. PMID: 35689015 Free PMC article.
-
Classification of particle trajectories in living cells: Machine learning versus statistical testing hypothesis for fractional anomalous diffusion.Phys Rev E. 2020 Sep;102(3-1):032402. doi: 10.1103/PhysRevE.102.032402. Phys Rev E. 2020. PMID: 33076015
-
A Jump-Distance-Based Parameter Inference Scheme for Particulate Trajectories.Biophys J. 2019 Jul 9;117(1):143-156. doi: 10.1016/j.bpj.2019.06.004. Epub 2019 Jun 12. Biophys J. 2019. PMID: 31235182 Free PMC article.
-
An Introduction to Infinite HMMs for Single-Molecule Data Analysis.Biophys J. 2017 May 23;112(10):2021-2029. doi: 10.1016/j.bpj.2017.04.027. Biophys J. 2017. PMID: 28538142 Free PMC article. Review.
-
Assessing Eating Behaviour Using Upper Limb Mounted Motion Sensors: A Systematic Review.Nutrients. 2019 May 24;11(5):1168. doi: 10.3390/nu11051168. Nutrients. 2019. PMID: 31137677 Free PMC article.
References
-
- Marchetti L., Bonsignore F., Gobbo F., Amodeo R., Calvello M., Jacob A., Signore G., Schirripa Spagnolo C., Porciani D., Mainardi M., et al. Fast-Diffusing P75 NTR Monomers Support Apoptosis and Growth Cone Collapse by Neurotrophin Ligands. Proc. Natl. Acad. Sci. USA. 2019;116:21563–21572. doi: 10.1073/pnas.1902790116. - DOI - PMC - PubMed
-
- Schirripa Spagnolo C., Moscardini A., Amodeo R., Beltram F., Luin S. Optimized Two-Color Single-Molecule Tracking of Fast-Diffusing Membrane Receptors. Adv. Opt. Mater. 2023;12:2302012. doi: 10.1002/adom.202302012. - DOI
Publication types
MeSH terms
Grants and funding
- RICBASE_2022_LUIN/Scuola Normale Superiore
- RICBASE_2023_LUIN/Scuola Normale Superiore
- National Quantum Science and Technology Institute (NQSTI - INVESTIMENTO 1.3; PE_00000023)/European Union Next-GenerationEU PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR - MISSIONE 4 COMPONENTE 2)
- Tuscany Health Ecosystem (THE - INVESTIMENTO 1.5; ECS_00000017)./European Union Next-GenerationEU PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR - MISSIONE 4 COMPONENTE 2)
LinkOut - more resources
Full Text Sources

