The Human OCTN Sub-Family: Gene and Protein Structure, Expression, and Regulation
- PMID: 39201429
- PMCID: PMC11354717
- DOI: 10.3390/ijms25168743
The Human OCTN Sub-Family: Gene and Protein Structure, Expression, and Regulation
Abstract
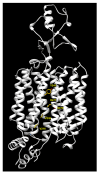
OCTN1 and OCTN2 are membrane transport proteins encoded by the SLC22A4 and SLC22A5 genes, respectively. Even though several transcripts have been predicted by bioinformatics for both genes, only one functional protein isoform has been described for each of them. Both proteins are ubiquitous, and depending on the physiopathological state of the cell, their expression is regulated by well-known transcription factors, although some aspects have been neglected. A plethora of missense variants with uncertain clinical significance are reported both in the dbSNP and the Catalogue of Somatic Mutations in Cancer (COSMIC) databases for both genes. Due to their involvement in human pathologies, such as inflammatory-based diseases (OCTN1/2), systemic primary carnitine deficiency (OCTN2), and drug disposition, it would be interesting to predict the impact of variants on human health from the perspective of precision medicine. Although the lack of a 3D structure for these two transport proteins hampers any speculation on the consequences of the polymorphisms, the already available 3D structures for other members of the SLC22 family may provide powerful tools to perform structure/function studies on WT and mutant proteins.
Keywords: IBD; OCTN1; OCTN2; SLC22 family; carnitine; gene expression and regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 2022JWT5XS/PRIN (Progetti di Ricerca di Interesse Nazionale), to L.P. granted by MUR (Ministry of University and Research) - Italy, funded by the European Union - Next Generation EU.
- PNC0000003/The National Plan for NRRP Complementary Investments - AdvaNced Technologies for Human-centrEd Medicine (ANTHEM) to M.G
LinkOut - more resources
Full Text Sources

