Phase Separation of SARS-CoV-2 Nucleocapsid Protein with TDP-43 Is Dependent on C-Terminus Domains
- PMID: 39201466
- PMCID: PMC11354357
- DOI: 10.3390/ijms25168779
Phase Separation of SARS-CoV-2 Nucleocapsid Protein with TDP-43 Is Dependent on C-Terminus Domains
Abstract
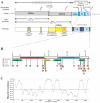
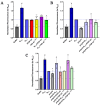
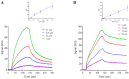
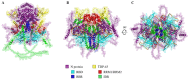
The SARS-CoV-2 nucleocapsid protein (N protein) is critical in viral replication by undergoing liquid-liquid phase separation to seed the formation of a ribonucleoprotein (RNP) complex to drive viral genomic RNA (gRNA) translation and in suppressing both stress granules and processing bodies, which is postulated to increase uncoated gRNA availability. The N protein can also form biomolecular condensates with a broad range of host endogenous proteins including RNA binding proteins (RBPs). Amongst these RBPs are proteins that are associated with pathological, neuronal, and glial cytoplasmic inclusions across several adult-onset neurodegenerative disorders, including TAR DNA binding protein 43 kDa (TDP-43) which forms pathological inclusions in over 95% of amyotrophic lateral sclerosis cases. In this study, we demonstrate that the N protein can form biomolecular condensates with TDP-43 and that this is dependent on the N protein C-terminus domain (N-CTD) and the intrinsically disordered C-terminus domain of TDP-43. This process is markedly accelerated in the presence of RNA. In silico modeling suggests that the biomolecular condensate that forms in the presence of RNA is composed of an N protein quadriplex in which the intrinsically disordered TDP-43 C terminus domain is incorporated.
Keywords: RNA binding proteins; amyotrophic lateral sclerosis; biomolecular condensates; neurodegeneration; nucleocapsid protein.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein.Nat Commun. 2021 Jan 21;12(1):502. doi: 10.1038/s41467-020-20768-y. Nat Commun. 2021. PMID: 33479198 Free PMC article.
-
SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs.EMBO J. 2020 Dec 15;39(24):e106478. doi: 10.15252/embj.2020106478. Epub 2020 Dec 4. EMBO J. 2020. PMID: 33200826 Free PMC article.
-
Phosphorylation in the Ser/Arg-rich region of the nucleocapsid of SARS-CoV-2 regulates phase separation by inhibiting self-association of a distant helix.J Biol Chem. 2024 Jun;300(6):107354. doi: 10.1016/j.jbc.2024.107354. Epub 2024 May 7. J Biol Chem. 2024. PMID: 38718862 Free PMC article.
-
Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization.J Biol Chem. 2024 Nov;300(11):107834. doi: 10.1016/j.jbc.2024.107834. Epub 2024 Sep 27. J Biol Chem. 2024. PMID: 39343000 Free PMC article. Review.
-
Phase separation and pathologic transitions of RNP condensates in neurons: implications for amyotrophic lateral sclerosis, frontotemporal dementia and other neurodegenerative disorders.Front Mol Neurosci. 2023 Sep 1;16:1242925. doi: 10.3389/fnmol.2023.1242925. eCollection 2023. Front Mol Neurosci. 2023. PMID: 37720552 Free PMC article. Review.
Cited by
-
The Role of TDP-43 in SARS-CoV-2-Related Neurodegenerative Changes.Viruses. 2025 May 19;17(5):724. doi: 10.3390/v17050724. Viruses. 2025. PMID: 40431734 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

