Structural Characterization of Heat Shock Protein 90β and Molecular Interactions with Geldanamycin and Ritonavir: A Computational Study
- PMID: 39201468
- PMCID: PMC11354266
- DOI: 10.3390/ijms25168782
Structural Characterization of Heat Shock Protein 90β and Molecular Interactions with Geldanamycin and Ritonavir: A Computational Study
Abstract
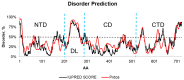
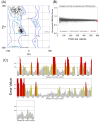
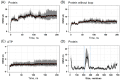
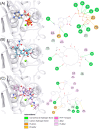
Drug repositioning is an important therapeutic strategy for treating breast cancer. Hsp90β chaperone is an attractive target for inhibiting cell progression. Its structure has a disordered and flexible linker region between the N-terminal and central domains. Geldanamycin was the first Hsp90β inhibitor to interact specifically at the N-terminal site. Owing to the toxicity of geldanamycin, we investigated the repositioning of ritonavir as an Hsp90β inhibitor, taking advantage of its proven efficacy against cancer. In this study, we used molecular modeling techniques to analyze the contribution of the Hsp90β linker region to the flexibility and interaction between the ligands geldanamycin, ritonavir, and Hsp90β. Our findings indicate that the linker region is responsible for the fluctuation and overall protein motion without disturbing the interaction between the inhibitors and the N-terminus. We also found that ritonavir established similar interactions with the substrate ATP triphosphate, filling the same pharmacophore zone.
Keywords: Hsp9β; cancer; chemical ligations; drug repurposing; geldanamycin; molecular docking; molecular dynamics; ritonavir.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

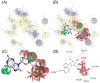
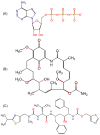
References
-
- Diniz C.S.G., Pellini A.C.G., Ribeiro A.G., Tedardi M.V., de Miranda M.J., Touso M.M., Baquero O.S., dos Santos P.C., Chiaravalloti-Neto F. Breast Cancer Mortality and Associated Factors in São Paulo State, Brazil: An Ecological Analysis. BMJ Open. 2017;7:e016395. doi: 10.1136/bmjopen-2017-016395. - DOI - PMC - PubMed
-
- Tilli T.M., Carels N., Tuszynski J.A., Pasdar M. Validation of a Network-Based Strategy for the Optimization of Combinatorial Target Selection in Breast Cancer Therapy: siRNA Knockdown of Network Targets in MDA-MB-231 Cells as an in Vitro Model for Inhibition of Tumor Development. Oncotarget. 2016;7:39. doi: 10.18632/oncotarget.11055. - DOI - PMC - PubMed
-
- Korsching E., Packeisen J., Liedtke C., Hungermann D., Wülfing P., van Diest P.J., Brandt B., Boecker W., Buerger H. The Origin of Vimentin Expression in Invasive Breast Cancer: Epithelial–Mesenchymal Transition, Myoepithelial Histogenesis or Histogenesis from Progenitor Cells with Bilinear Differentiation Potential? J. Pathol. 2005;206:451–457. doi: 10.1002/path.1797. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

