Repetitive Sequence Stability in Embryonic Stem Cells
- PMID: 39201503
- PMCID: PMC11354519
- DOI: 10.3390/ijms25168819
Repetitive Sequence Stability in Embryonic Stem Cells
Abstract
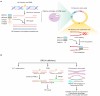
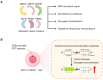
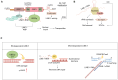
Repetitive sequences play an indispensable role in gene expression, transcriptional regulation, and chromosome arrangements through trans and cis regulation. In this review, focusing on recent advances, we summarize the epigenetic regulatory mechanisms of repetitive sequences in embryonic stem cells. We aim to bridge the knowledge gap by discussing DNA damage repair pathway choices on repetitive sequences and summarizing the significance of chromatin organization on repetitive sequences in response to DNA damage. By consolidating these insights, we underscore the critical relationship between the stability of repetitive sequences and early embryonic development, seeking to provide a deeper understanding of repetitive sequence stability and setting the stage for further research and potential therapeutic strategies in developmental biology and regenerative medicine.
Keywords: DNA damage; embryonic stem cells; epigenetic regulation; repetitive sequences.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Hoyt S.J., Storer J.M., Hartley G.A., Grady P.G.S., Gershman A., de Lima L.G., Limouse C., Halabian R., Wojenski L., Rodriguez M., et al. From telomere to telomere: The transcriptional and epigenetic state of human repeat elements. Science. 2022;376:eabk3112. doi: 10.1126/science.abk3112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

