Wastewater Surveillance of SARS-CoV-2 in Zambia: An Early Warning Tool
- PMID: 39201525
- PMCID: PMC11354861
- DOI: 10.3390/ijms25168839
Wastewater Surveillance of SARS-CoV-2 in Zambia: An Early Warning Tool
Abstract
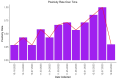
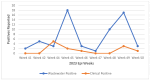
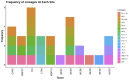
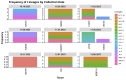
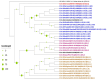
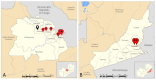
Wastewater-based surveillance has emerged as an important method for monitoring the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). This study investigated the presence of SARS-CoV-2 in wastewater in Zambia. We conducted a longitudinal study in the Copperbelt and Eastern provinces of Zambia from October 2023 to December 2023 during which 155 wastewater samples were collected. The samples were subjected to three different concentration methods, namely bag-mediated filtration, skimmed milk flocculation, and polythene glycol-based concentration assays. Molecular detection of SARS-CoV-2 nucleic acid was conducted using real-time Polymerase Chain Reaction (PCR). Whole genome sequencing was conducted using Illumina COVIDSEQ assay. Of the 155 wastewater samples, 62 (40%) tested positive for SARS-CoV-2. Of these, 13 sequences of sufficient length to determine SARS-CoV-2 lineages were obtained and 2 sequences were phylogenetically analyzed. Various Omicron subvariants were detected in wastewater including BA.5, XBB.1.45, BA.2.86, and JN.1. Some of these subvariants have been detected in clinical cases in Zambia. Interestingly, phylogenetic analysis positioned a sequence from the Copperbelt Province in the B.1.1.529 clade, suggesting that earlier Omicron variants detected in late 2021 could still be circulating and may not have been wholly replaced by newer subvariants. This study stresses the need for integrating wastewater surveillance of SARS-CoV-2 into mainstream strategies for monitoring SARS-CoV-2 circulation in Zambia.
Keywords: COVID-19; SARS-CoV-2; Zambia; early warning; surveillance; wastewater.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genomic Surveillance of SARS-CoV-2 in the Southern Province of Zambia: Detection and Characterization of Alpha, Beta, Delta, and Omicron Variants of Concern.Viruses. 2022 Aug 24;14(9):1865. doi: 10.3390/v14091865. Viruses. 2022. PMID: 36146671 Free PMC article.
-
Detection and Characterisation of SARS-CoV-2 in Eastern Province of Zambia: A Retrospective Genomic Surveillance Study.Int J Mol Sci. 2024 Jun 7;25(12):6338. doi: 10.3390/ijms25126338. Int J Mol Sci. 2024. PMID: 38928045 Free PMC article.
-
Rapid spread of the SARS-CoV-2 Omicron XDR lineage derived from recombination between XBB and BA.2.86 subvariants circulating in Brazil in late 2023.Microbiol Spectr. 2025 Jan 7;13(1):e0119324. doi: 10.1128/spectrum.01193-24. Epub 2024 Nov 29. Microbiol Spectr. 2025. PMID: 39611827 Free PMC article.
-
Wastewater Genomic Surveillance Captures Early Detection of Omicron in Utah.Microbiol Spectr. 2023 Jun 15;11(3):e0039123. doi: 10.1128/spectrum.00391-23. Epub 2023 May 8. Microbiol Spectr. 2023. PMID: 37154725 Free PMC article.
-
Detection of SARS-CoV-2 B.1.351 (Beta) Variant through Wastewater Surveillance before Case Detection in a Community, Oregon, USA.Emerg Infect Dis. 2022 Jun;28(6):1101-1109. doi: 10.3201/eid2806.211821. Epub 2022 Apr 22. Emerg Infect Dis. 2022. PMID: 35452383 Free PMC article. Review.
Cited by
-
Dynamics of SARS-CoV-2 Mutations in Wastewater Provide Insights into the Circulation of Virus Variants in the Population.Int J Mol Sci. 2025 May 1;26(9):4324. doi: 10.3390/ijms26094324. Int J Mol Sci. 2025. PMID: 40362561 Free PMC article.
References
-
- Denaro M., Ferro E., Barrano G., Meli S., Busacca M., Corallo D., Capici A., Zisa A., Cucuzza L., Gradante S., et al. Monitoring of SARS-CoV-2 Infection in Ragusa Area: Next Generation Sequencing and Serological Analysis. Int. J. Mol. Sci. 2023;24:4742. doi: 10.3390/ijms24054742. - DOI - PMC - PubMed
-
- Carabelli A.M., Peacock T.P., Thorne L.G., Harvey W.T., Hughes J., de Silva T.I., Peacock S.J., Barclay W.S., de Silva T.I., Towers G.J., et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023;21:162–177. doi: 10.1038/s41579-022-00841-7. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

