Evolution and Competitive Struggles of Lactiplantibacillus plantarum under Different Oxygen Contents
- PMID: 39201547
- PMCID: PMC11354895
- DOI: 10.3390/ijms25168861
Evolution and Competitive Struggles of Lactiplantibacillus plantarum under Different Oxygen Contents
Abstract
Lactiplantibacillus (Lb.) plantarum is known as a benign bacterium found in various habitats, including the intestines of animals and fermented foods. Since animal intestines lack oxygen, while fermented foods provide a limited or more oxygen environment, this study aimed to investigate whether there were genetic differences in the growth of Lb. plantarum under aerobic vs. anaerobic conditions. Genomic analysis of Lb. plantarum obtained from five sources-animals, dairy products, fermented meat, fermented vegetables, and humans-was conducted. The analysis included not only an examination of oxygen-utilizing genes but also a comparative pan-genomic analysis to investigate evolutionary relationships between genomes. The ancestral gene analysis of the evolutionary pathway classified Lb. plantarum into groups A and B, with group A further subdivided into A1 and A2. It was confirmed that group A1 does not possess the narGHIJ operon, which is necessary for energy production under limited oxygen conditions. Additionally, it was found that group A1 has experienced more gene acquisition and loss compared to groups A2 and B. Despite an initial assumption that there would be genetic distinctions based on the origin (aerobic or anaerobic conditions), it was observed that such differentiation could not be attributed to the origin. However, the evolutionary process indicated that the loss of genes related to nitrate metabolism was essential in anaerobic or limited oxygen conditions, contrary to the initial hypothesis.
Keywords: Lactiplantibacillus plantarum; core-genome; nitrate metabolism; pan-genome.
Conflict of interest statement
Author Moon-Hee Sung is employed by the company KookminBio Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
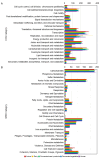
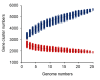

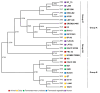


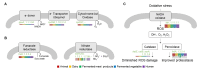
References
-
- Imlay J.A. How oxygen damages microbes: Oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 2002;46:111–153. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

