Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.)
- PMID: 39201587
- PMCID: PMC11354645
- DOI: 10.3390/ijms25168901
Transcriptome Profiles Reveal Key Regulatory Networks during Single and Multifactorial Stresses Coupled with Melatonin Treatment in Pitaya (Selenicereus undatus L.)
Abstract
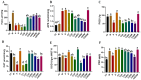
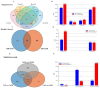
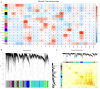
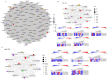
In response to evolving climatic conditions, plants frequently confront multiple abiotic stresses, necessitating robust adaptive mechanisms. This study focuses on the responses of Selenicereus undatus L. to both individual stresses (cadmium; Cd, salt; S, and drought; D) and their combined applications, with an emphasis on evaluating the mitigating effects of (M) melatonin. Through transcriptome analysis, this study identifies significant gene expression changes and regulatory network activations. The results show that stress decreases pitaya growth rates by 30%, reduces stem and cladode development by 40%, and increases Cd uptake under single and combined stresses by 50% and 70%, respectively. Under stress conditions, enhanced activities of H2O2, POD, CAT, APX, and SOD and elevated proline content indicate strong antioxidant defenses. We identified 141 common DEGs related to stress tolerance, most of which were related to AtCBP, ALA, and CBP pathways. Interestingly, the production of genes related to signal transduction and hormones, including abscisic acid and auxin, was also significantly induced. Several calcium-dependent protein kinase genes were regulated during M and stress treatments. Functional enrichment analysis showed that most of the DEGs were enriched during metabolism, MAPK signaling, and photosynthesis. In addition, weighted gene co-expression network analysis (WGCNA) identified critical transcription factors (WRKYs, MYBs, bZIPs, bHLHs, and NACs) associated with antioxidant activities, particularly within the salmon module. This study provides morpho-physiological and transcriptome insights into pitaya's stress responses and suggests molecular breeding techniques with which to enhance plant resistance.
Keywords: Selenicereus undatus L.; TFs; WGCNA; single and combined stresses; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Comparative Physiological and Transcriptomics Profiling Provides Integrated Insight into Melatonin Mediated Salt and Copper Stress Tolerance in Selenicereus undatus L.Plants (Basel). 2024 Dec 23;13(24):3602. doi: 10.3390/plants13243602. Plants (Basel). 2024. PMID: 39771301 Free PMC article.
-
Isolation and characterization of a catalase gene "HuCAT3" from pitaya (Hylocereus undatus) and its expression under abiotic stress.Gene. 2015 May 25;563(1):63-71. doi: 10.1016/j.gene.2015.03.007. Epub 2015 Mar 6. Gene. 2015. PMID: 25752288
-
Unique regulatory network of dragon fruit simultaneously mitigates the effect of vanadium pollutant and environmental factors.Physiol Plant. 2024 Jul-Aug;176(4):e14416. doi: 10.1111/ppl.14416. Physiol Plant. 2024. PMID: 38952344
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize.Biotechnol Lett. 2022 Mar;44(3):367-386. doi: 10.1007/s10529-022-03221-6. Epub 2022 Mar 16. Biotechnol Lett. 2022. PMID: 35294695 Review.
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

