Different Prostatic Tissue Microbiomes between High- and Low-Grade Prostate Cancer Pathogenesis
- PMID: 39201629
- PMCID: PMC11354394
- DOI: 10.3390/ijms25168943
Different Prostatic Tissue Microbiomes between High- and Low-Grade Prostate Cancer Pathogenesis
Abstract
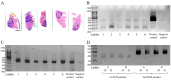
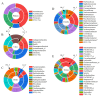
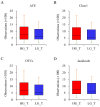
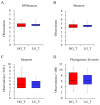
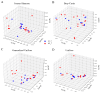
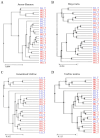
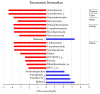
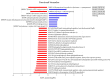
Numerous human pathologies, such as neoplasia, are related to particular bacteria and changes in microbiome constituents. To investigate the association between an imbalance of bacteria and prostate carcinoma, the microbiome and gene functionality from tissues of patients with high-grade prostate tumor (HGT) and low-grade prostate tumor (LGT) were compared utilizing next-generation sequencing (NGS) technology. The results showed abnormalities in the bacterial profiles between the HGT and LGT specimens, indicating alterations in the make-up of bacterial populations and gene functionalities. The HGT specimens showed higher frequencies of Cutibacterium, Pelomonas, and Corynebacterium genera than the LGT specimens. Cell proliferation and cytokine assays also showed a significant proliferation of prostate cancer cells and elevated cytokine levels in the cells treated with Cutibacterium, respectively, supporting earlier findings. In summary, the HGT and LGT specimens showed differences in bacterial populations, suggesting that different bacterial populations might characterize high-grade and low-grade prostate malignancies.
Keywords: Cutibacterium; biomarker; high-grade prostate tumor (HGT); low-grade prostate tumor (LGT); prostate cancer; therapeutic targets.
Conflict of interest statement
The authors have no conflicts of interest relevant to this study to disclose.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

