Genome-Wide Identification, Expression, and Protein Analysis of CKX and IPT Gene Families in Radish (Raphanus sativus L.) Reveal Their Involvement in Clubroot Resistance
- PMID: 39201660
- PMCID: PMC11354997
- DOI: 10.3390/ijms25168974
Genome-Wide Identification, Expression, and Protein Analysis of CKX and IPT Gene Families in Radish (Raphanus sativus L.) Reveal Their Involvement in Clubroot Resistance
Abstract
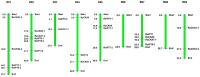
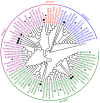
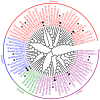
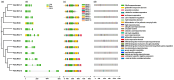
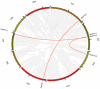
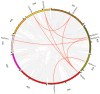
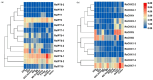
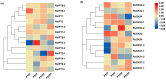
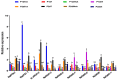
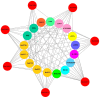
Cytokinins (CKs) are a group of phytohormones that are involved in plant growth, development, and disease resistance. The isopentenyl transferase (IPT) and cytokinin oxidase/dehydrogenase (CKX) families comprise key enzymes controlling CK biosynthesis and degradation. However, an integrated analysis of these two gene families in radish has not yet been explored. In this study, 13 RsIPT and 12 RsCKX genes were identified and characterized, most of which had four copies in Brassica napus and two copies in radish and other diploid Brassica species. Promoter analysis indicated that the genes contained at least one phytohormone or defense and stress responsiveness cis-acting element. RsIPTs and RsCKXs were expanded through segmental duplication. Moreover, strong purifying selection drove the evolution of the two gene families. The expression of the RsIPT and RsCKX genes distinctly showed diversity in different tissues and developmental stages of the root. Expression profiling showed that RsCKX1-1/1-2/1-3 was significantly upregulated in club-resistant materials during primary infection, suggesting their vital function in clubroot resistance. The interaction network of CKX proteins with similar 3D structures also reflected the important role of RsCKX genes in disease resistance. This study provides a foundation for further functional study on the IPT and CKX genes for clubroot resistance improvement in Raphanus.
Keywords: 3D structure; Raphanus sativus; RsCKX; RsIPT; clubroot resistance; expression profile; interaction network.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Hirai M. Genetic analysis of clubroot resistance in brassica crops. Bred. Sci. 2006;56:223–229. doi: 10.1270/jsbbs.56.223. - DOI
-
- Wang J., Huang Y.L., Li X.L., Li H.Z. Research progress in clubroot of crucifers. Plant Prot. 2011;37:153–158.
-
- Yang H., Yuan Y., Wei X., Zhang X., Wang H., Song J., Li X. A new identification method reveals the resistance of an extensive-source radish collection to Plasmodiophora brassicae race 4. Agronomy. 2021;11:792. doi: 10.3390/agronomy11040792. - DOI
-
- Ingram D.S., Tommerup I.C., Ingram D.S., Brian P.W. The life history of Plasmodiophora bracssicae Woron. Proc. B Soc. Lond. B. 1972;180:103–112.
-
- Wallenhammar A.C. Prevalence of Plasmodiophora brassicae in a spring oilseed rape growing area in central sweden and factors influencing soil infestation levels. Plant Pathol. 2010;45:710–719. doi: 10.1046/j.1365-3059.1996.d01-173.x. - DOI
MeSH terms
Substances
Grants and funding
- 2016YFD0100204-02/National Key Research and Development Program of China
- CAAS-ASTIP-IVFCAAS/Science and Technology Innovation Pro-gram of the Chinese Academy of Agricultural Sciences
- CAAS-XTCX2016001/Science and Technology Innovation Pro-gram of the Chinese Academy of Agricultural Sciences
- CAAS-XTCX2016016-4-4/Science and Technology Innovation Pro-gram of the Chinese Academy of Agricultural Sciences
- 31772303/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials

