Genome-Wide Analyses of CCHC Family Genes and Their Expression Profiles under Drought Stress in Rose (Rosa chinensis)
- PMID: 39201669
- PMCID: PMC11354476
- DOI: 10.3390/ijms25168983
Genome-Wide Analyses of CCHC Family Genes and Their Expression Profiles under Drought Stress in Rose (Rosa chinensis)
Abstract
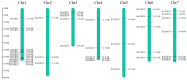
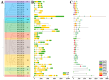
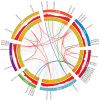
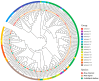
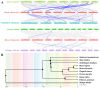
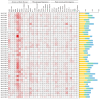
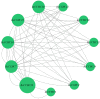
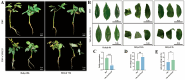
CCHC-type zinc finger proteins (CCHC-ZFPs), ubiquitous across plant species, are integral to their growth, development, hormonal regulation, and stress adaptation. Roses (Rosa sp.), as one of the most significant and extensively cultivated ornamentals, account for more than 30% of the global cut-flower market. Despite its significance, the CCHC gene family in roses (Rosa sp.) remains unexplored. This investigation identified and categorized 41 CCHC gene members located on seven chromosomes of rose into 14 subfamilies through motif distribution and phylogenetic analyses involving ten additional plant species, including Ginkgo biloba, Ostreococcus lucimarinus, Arabidopsis thaliana, and others. This study revealed that dispersed duplication likely plays a crucial role in the diversification of the CCHC genes, with the Ka/Ks ratio suggesting a history of strong purifying selection. Promoter analysis highlighted a rich presence of cis-acting regulatory elements linked to both abiotic and biotic stress responses. Differential expression analysis under drought conditions grouped the 41 CCHC gene members into five distinct clusters, with those in group 4 exhibiting pronounced regulation in roots and leaves under severe drought. Furthermore, virus-induced gene silencing (VIGS) of the RcCCHC25 member from group 4 compromised drought resilience in rose foliage. This comprehensive analysis lays the groundwork for further investigations into the functional dynamics of the CCHC gene family in rose physiology and stress responses.
Keywords: VIGS; abiotic stress; cut flowers; gene family analysis; zinc finger proteins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Drought Stress in Roses: A Comprehensive Review of Morphophysiological, Biochemical, and Molecular Responses.Int J Mol Sci. 2025 Apr 30;26(9):4272. doi: 10.3390/ijms26094272. Int J Mol Sci. 2025. PMID: 40362508 Free PMC article. Review.
-
Genome-Wide Identification and Expression Analysis of the AP2/ERF Transcription Factor Gene Family in Hybrid Tea Rose Under Drought Stress.Int J Mol Sci. 2024 Nov 29;25(23):12849. doi: 10.3390/ijms252312849. Int J Mol Sci. 2024. PMID: 39684560 Free PMC article.
-
Genome-wide analysis of BURP genes and identification of a BURP-V gene RcBURP4 in Rosa chinensis.Plant Cell Rep. 2022 Feb;41(2):395-413. doi: 10.1007/s00299-021-02815-0. Epub 2021 Nov 24. Plant Cell Rep. 2022. PMID: 34820714
-
Genome-wide analysis of the rose (Rosa chinensis) NAC family and characterization of RcNAC091.Plant Mol Biol. 2022 Apr;108(6):605-619. doi: 10.1007/s11103-022-01250-3. Epub 2022 Feb 15. Plant Mol Biol. 2022. PMID: 35169911
-
CCCH-type zinc finger family in maize: genome-wide identification, classification and expression profiling under abscisic acid and drought treatments.PLoS One. 2012;7(7):e40120. doi: 10.1371/journal.pone.0040120. Epub 2012 Jul 6. PLoS One. 2012. PMID: 22792223 Free PMC article.
Cited by
-
Drought Stress in Roses: A Comprehensive Review of Morphophysiological, Biochemical, and Molecular Responses.Int J Mol Sci. 2025 Apr 30;26(9):4272. doi: 10.3390/ijms26094272. Int J Mol Sci. 2025. PMID: 40362508 Free PMC article. Review.
References
-
- Shi L., Kim W.S. Shoot growth and physiological dis-order of cut rose ‘charming black’ as affected by drought stress during nocturnal supplemental lighting. Hortic. Environ. Biotechnol. 2014;55:91–96. doi: 10.1007/s13580-014-0166-7. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

