Deciphering the Plastome and Molecular Identities of Six Medicinal "Doukou" Species
- PMID: 39201691
- PMCID: PMC11354342
- DOI: 10.3390/ijms25169005
Deciphering the Plastome and Molecular Identities of Six Medicinal "Doukou" Species
Abstract
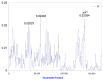
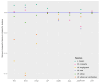
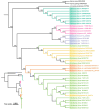
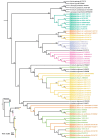
The genus Amomum includes over 111 species, 6 of which are widely utilized as medicinal plants and have already undergone taxonomic revision. Due to their morphological similarities, the presence of counterfeit and substandard products remains a challenge. Accurate plant identification is, therefore, essential to address these issues. This study utilized 11 newly sequenced samples and extensive NCBI data to perform molecular identification of the six medicinal "Doukou" species. The plastomes of these species exhibited a typical quadripartite structure with a conserved gene content. However, independent variation shifts of the SC/IR boundaries existed between and within species. The comprehensive set of genetic sequences, including ITS, ITS1, ITS2, complete plastomes, matK, rbcL, psbA-trnH, and ycf1, showed varying discrimination of the six "Doukou" species based on both distance and phylogenetic tree methods. Among these, the ITS, ITS1, and complete plastome sequences demonstrated the highest identification success rate (3/6), followed by ycf1 (2/6), and then ITS2, matK, and psbA-trnH (1/6). In contrast, rbcL failed to identify any species. This research established a basis for a reliable molecular identification method for medicinal "Doukou" plants to protect wild plant resources, promote the sustainable use of medicinal plants, and restrict the exploitation of these resources.
Keywords: DNA barcoding; ITS; medicinal plants; plastome; species identification; “Doukou”.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Gotelli N.J., Colwell R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001;4:379–391. doi: 10.1046/j.1461-0248.2001.00230.x. - DOI
-
- Soulé M.E., Wilcox B.A. Conservation Biology. An Evolutionary-Ecological Perspective. Addison-Wesle; London, UK: 1980. p. 395.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

