Wuhan Sequence-Based Recombinant Antigens Expressed in E. coli Elicit Antibodies Capable of Binding with Omicron S-Protein
- PMID: 39201702
- PMCID: PMC11354337
- DOI: 10.3390/ijms25169016
Wuhan Sequence-Based Recombinant Antigens Expressed in E. coli Elicit Antibodies Capable of Binding with Omicron S-Protein
Abstract
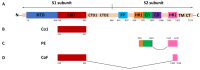
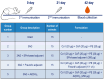
The development of cross-reactive vaccines is one of the central aims of modern vaccinology. Continuous mutation and the emergence of new SARS-CoV-2 variants and subvariants create the problem of universal coronavirus vaccine design. Previously, the authors devised three recombinant coronavirus antigens, which were based on the sequence collected in 2019 (the Wuhan variant) and produced in an E. coli bacterial expression system. The present work has shown, for the first time, that these recombinant antigens induce the production of antibodies that clearly interact with produced in CHO full-length S-protein of the Omicron variant. The immunogenicity of these recombinant antigens was studied in formulations with different adjuvants: Freund's adjuvant, Al(OH)3 and an adjuvant based on spherical particles (SPs), which are structurally modified plant virus. All adjuvanted formulations effectively stimulated Omicron-specific IgG production in mice. These universal coronavirus antigens could be considered the main component for the further development of broad-spectrum coronavirus vaccines for the prevention of SARS-CoV-2 infection. The present work also provides evidence that the synthetic biology approach is a promising strategy for the development of highly cross-reactive vaccines. Moreover, it is important to note that the bacterial expression system might be appropriate for the production of antigenically active universal antigens.
Keywords: Omicron variant; SARS-CoV-2; plant virus-based adjuvant; recombinant protein vaccine.
Conflict of interest statement
The authors declare that this study received funding from the Russian Science foundation (Grant number 24-14-00032) and R-Pharm (providing of SARS-CoV-2 commercial proteins of Omicron). The funders were not involved in the study design; the collection, analysis or interpretation of the data; the writing of this article or the decision to submit it for publication.
Figures


Similar articles
-
Mammalian cell expressed recombinant trimeric spike protein is a potent vaccine antigen and confers near-complete protection against SARS-CoV-2 infection in Hamster.Vaccine. 2024 Aug 13;42(20):126099. doi: 10.1016/j.vaccine.2024.06.066. Epub 2024 Jul 8. Vaccine. 2024. PMID: 38981743
-
SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen.J Med Virol. 2021 Feb;93(2):892-898. doi: 10.1002/jmv.26320. Epub 2020 Oct 5. J Med Virol. 2021. PMID: 32691875 Free PMC article.
-
SARS-CoV-2 spike-based virus-like particles incorporate influenza H1/N1 antigens and induce dual immunity in mice.Vaccine. 2024 Dec 2;42(26):126463. doi: 10.1016/j.vaccine.2024.126463. Epub 2024 Oct 30. Vaccine. 2024. PMID: 39481241
-
The Development of Epitope-Based Recombinant Protein Vaccines against SARS-CoV-2.AAPS J. 2024 Aug 13;26(5):93. doi: 10.1208/s12248-024-00963-1. AAPS J. 2024. PMID: 39138686 Review.
-
Severe acute respiratory syndrome-coronavirus-2 spike (S) protein based vaccine candidates: State of the art and future prospects.Rev Med Virol. 2021 May;31(3):e2183. doi: 10.1002/rmv.2183. Epub 2020 Oct 15. Rev Med Virol. 2021. PMID: 33594794 Free PMC article. Review.
Cited by
-
Adaptation of the Vaccine Prophylaxis Strategy to Variants of the SARS-CoV-2 Virus.Vaccines (Basel). 2025 Jul 17;13(7):761. doi: 10.3390/vaccines13070761. Vaccines (Basel). 2025. PMID: 40733738 Free PMC article. Review.
References
-
- Rabaan A.A., Al-Ahmed S.H., Haque S., Sah R., Tiwari R., Malik Y.S., Dhama K., Yatoo M.I., Bonilla-Aldana D.K., Rodriguez-Morales A.J. SARS-CoV-2, SARS-CoV, and MERS-COV: A comparative overview. Infez. Med. 2020;28:174–184. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

