A Fungicide, Fludioxonil, Formed the Polyploid Giant Cancer Cells and Induced Metastasis and Stemness in MDA-MB-231 Triple-Negative Breast Cancer Cells
- PMID: 39201710
- PMCID: PMC11354328
- DOI: 10.3390/ijms25169024
A Fungicide, Fludioxonil, Formed the Polyploid Giant Cancer Cells and Induced Metastasis and Stemness in MDA-MB-231 Triple-Negative Breast Cancer Cells
Abstract
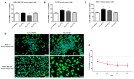
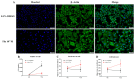
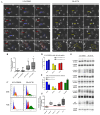
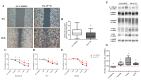
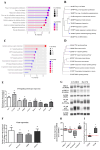
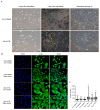
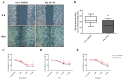
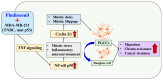
Fludioxonil, an antifungal agent used as a pesticide, leaves a measurable residue in fruits and vegetables. It has been identified to cause endocrine disruption, interrupt normal development, and cause various diseases such as cancers. In this study, fludioxonil was examined for its effects on the development and metastasis of breast cancer cells. On fludioxonil exposure (10-5 M) for 72 h, mutant p53 (mutp53) MDA-MB-231 triple-negative breast cancer (TNBC) cells significantly inhibited cell viability and developed into polyploid giant cancer cells (PGCCs), with an increase in the number of nuclei and expansion in the cell body size. Fludioxonil exposure disrupted the normal cell cycle phase ratio, resulting in a new peak. In addition, PGCCs showed greater motility than the control and were resistant to anticancer drugs, i.e., doxorubicin, cisplatin, and 5-fluorouracil. Cyclin E1, nuclear factor kappa B (NF-κB), and p53 expressions were remarkably increased, and the expression of cell cycle-, epithelial-mesenchymal-transition (EMT)-, and cancer stemness-related proteins were increased in the PGCCs. The daughter cells obtained from PGCCs had the single nucleus but maintained their enlarged cell size and showed greater cell migration ability and resistance to the anticancer agents. Consequently, fludioxonil accumulated Cyclin E1 and promoted the inflammatory cytokine-enriched microenvironment through the up-regulation of TNF and NF-κB which led to the transformation to PGCCs via abnormal cell cycles such as mitotic delay and mitotic slippage in mutp53 TNBC MDA-MB-231 cells. PGCCs and their daughter cells exhibited significant migration ability, chemo-resistance, and cancer stemness. These results strongly suggest that fludioxonil, as an inducer of potential genotoxicity, may induce the formation of PGCCs, leading to the formation of metastatic and stem cell-like breast cancer cells.
Keywords: TNBC; fludioxonil; fungicide; polyploid giant cancer cell.
Conflict of interest statement
The authors do not have any conflicts of interest to declare.
Figures

References
-
- Erenpreisa J., Ivanov A., Wheatley S.P., Kosmacek E.A., Ianzini F., Anisimov A.P., Mackey M., Davis P.J., Plakhins G., Illidge T.M. Endopolyploidy in irradiated p53-deficient tumour cell lines: Persistence of cell division activity in giant cells expressing Aurora-B kinase. Cell Biol. Int. 2008;32:1044–1056. doi: 10.1016/j.cellbi.2008.06.003. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

