Genome-Wide Characterization of the BTB Gene Family in Poplar and Expression Analysis in Response to Hormones and Biotic/Abiotic Stresses
- PMID: 39201733
- PMCID: PMC11354360
- DOI: 10.3390/ijms25169048
Genome-Wide Characterization of the BTB Gene Family in Poplar and Expression Analysis in Response to Hormones and Biotic/Abiotic Stresses
Abstract
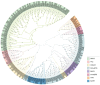
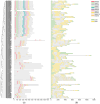
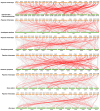
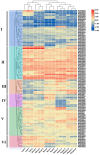
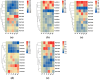
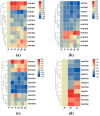
The BTB (Broad-complex, tramtrack, and bric-a-brac) gene family, characterized by a highly conserved BTB domain, is implicated in a spectrum of biological processes, encompassing growth and development, as well as stress responses. Characterization and functional studies of BTB genes in poplar are still limited, especially regarding their response to hormones and biotic/abiotic stresses. In this study, we conducted an HMMER search in conjunction with BLASTp and identified 95 BTB gene models in Populus trichocarpa. Through domain motif and phylogenetic relationship analyses, these proteins were classified into eight families, NPH3, TAZ, Ankyrin, only BTB, BACK, Armadillo, TPR, and MATH. Collinearity analysis of poplar BTB genes with homologs in six other species elucidated evolutionary relationships and functional conservations. RNA-seq analysis of five tissues of poplar identified BTB genes as playing a pivotal role during developmental processes. Comprehensive RT-qPCR analysis of 11 BTB genes across leaves, roots, and xylem tissues revealed their responsive expression patterns under diverse hormonal and biotic/abiotic stress conditions, with varying degrees of regulation observed in the results. This study marks the first in-depth exploration of the BTB gene family in poplar, providing insights into the potential roles of BTB genes in hormonal regulation and response to stress.
Keywords: BTB gene family; Populus trichocarpa; biotic/abiotic stresses; expression patterns; hormones treatment.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

