Characterization and Functional Analysis of the 17-Beta Hydroxysteroid Dehydrogenase 2 (hsd17b2) Gene during Sex Reversal in the Ricefield Eel (Monopterus albus)
- PMID: 39201749
- PMCID: PMC11354438
- DOI: 10.3390/ijms25169063
Characterization and Functional Analysis of the 17-Beta Hydroxysteroid Dehydrogenase 2 (hsd17b2) Gene during Sex Reversal in the Ricefield Eel (Monopterus albus)
Abstract
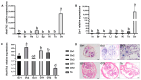
In mammals, 17-beta hydroxysteroid dehydrogenase 2 (Hsd17b2) enzyme specifically catalyzes the oxidation of the C17 hydroxyl group and efficiently regulates the activities of estrogens and androgens to prevent diseases induced by hormone disorders. However, the functions of the hsd17b2 gene involved in animal sex differentiation are still largely unclear. The ricefield eel (Monopterus albus), a protogynous hermaphroditic fish with a small genome size (2n = 24), is usually used as an ideal model to study the mechanism of sex differentiation in vertebrates. Therefore, in this study, hsd17b2 gene cDNA was cloned and its mRNA expression profiles were determined in the ricefield eel. The cloned cDNA fragment of hsd17b2 was 1230 bp, including an open reading frame of 1107 bp, encoding 368 amino acid residues with conserved catalytic subunits. Moreover, real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis showed that hsd17b2 mRNA expressed strongly in the ovaries at early developmental stages, weakly in liver and intestine, and barely in testis and other tissues. In particular, hsd17b2 mRNA expression was found to peak in ovaries of young fish and ovotestis at the early stage, and eventually declined in gonads from the late ovotestis to testis. Likewise, chemical in situ hybridization results indicated that the hsd17b2 mRNA signals were primarily detected in the cytoplasm of oogonia and oocytes at stage I-II, subsequently concentrated in the granulosa cells around the oocytes at stage Ⅲ-Ⅳ, but undetectable in mature oocytes and male germ cells. Intriguingly, in ricefield eel ovaries, hsd17b2 mRNA expression could be significantly reduced by 17β-estradiol (E2) or tamoxifen (17β-estradiol inhibitor, E2I) induction at a low concentration (10 ng/mL) and increased by E2I induction at a high concentration (100 ng/mL). On the other hand, both the melatonin (MT) and flutamide (androgen inhibitor, AI) induction could significantly decrease hsd17b2 mRNA expression in the ovary of ricefield eel. This study provides a clue for demonstrating the mechanism of sexual differentiation in fish. The findings of our study imply that the hsd17b2 gene could be a key regulator in sexual differentiation and modulate sex reversal in the ricefield eel and other hermaphroditic fishes.
Keywords: chemical in situ hybridization; hormone induction; hsd17b2; ricefield eel; sex reversal.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





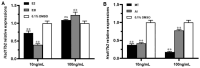
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

