Pitfalls When Determining HNA-1 Genotypes and Finding Novel Alleles
- PMID: 39201813
- PMCID: PMC11354314
- DOI: 10.3390/ijms25169127
Pitfalls When Determining HNA-1 Genotypes and Finding Novel Alleles
Abstract
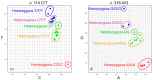
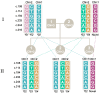
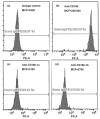
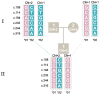
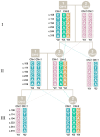
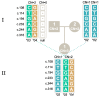
Genetic variation in the FCGR3B gene is responsible for different variants of human neutrophil antigen 1 (HNA-1). Laboratory techniques currently utilized for routine HNA-1 genotyping, predominantly PCR-sequence-specific primer (PCR-SSP) and PCR-sequence-based typing (PCR-SBT), lack specificity for FCGR3B. This study compares the capabilities and limitations of existing technologies including an in-house TaqMan PCR, a commercial PCR-SSP test, PCR-SBT and multiplex ligation-dependent probe amplification (MLPA) with those of a long-read nanopore sequencing assay. Testing was performed with both related and unrelated Danish samples with different copy numbers and/or rare alleles. Long-read nanopore sequencing was validated by blind testing of ten English samples. The results showed that FCGR3B copy numbers correlate with a dose-dependent distribution of alleles that complicates genotyping by TaqMan PCR, PCR-SSP and PCR-SBT, due to co-amplification of the homologous FCGR3A gene. MLPA can correctly quantify the dose-dependent distribution but not detect novel variants. Long-read nanopore sequencing showed high specificity for FCGR3B and was able to detect dosage-dependent distribution, and rare and novel variants that were previously not described. Current HNA-1 genotyping methods cannot produce unambiguous allele-level results, whereas long-read nanopore sequencing has shown the potential to resolve observed ambiguities, identify new HNA-1 variants and allow definitive allele assignment.
Keywords: FCGR3A; FCGR3B; HNA-1; PCR; copy number variation; genotyping; long-read sequencing; nanopore.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genotyping of human neutrophil antigens by polymerase chain reaction sequence-based typing.Blood Transfus. 2014 Jan;12 Suppl 1(Suppl 1):s292-8. doi: 10.2450/2013.0308-12. Epub 2013 Jun 19. Blood Transfus. 2014. PMID: 23867183 Free PMC article.
-
HNA-1, -3, -4, and -5 genotyping using multiplex PCR among southern Thais: Developing continual HNA-1 null detection.J Clin Lab Anal. 2019 Jan;33(1):e22651. doi: 10.1002/jcla.22651. Epub 2018 Aug 13. J Clin Lab Anal. 2019. PMID: 30105756 Free PMC article.
-
Frequencies of HNA-1, HNA-3, HNA-4, and HNA-5 in the Danish and Zambian populations determined using a novel TaqMan real time polymerase chain reaction method.Tissue Antigens. 2012 Sep;80(3):249-53. doi: 10.1111/j.1399-0039.2012.01912.x. Epub 2012 Jun 15. Tissue Antigens. 2012. PMID: 22703110
-
Genotyping of human neutrophil antigens 1, 3, 4 and 5 using a novel multiplex polymerase chain reaction.Transfus Med. 2019 Apr;29(2):110-115. doi: 10.1111/tme.12594. Transfus Med. 2019. PMID: 30974499
-
Human neutrophil antigens: Nature, clinical significance and detection.Int J Immunogenet. 2021 Apr;48(2):145-156. doi: 10.1111/iji.12514. Epub 2020 Sep 24. Int J Immunogenet. 2021. PMID: 32970372 Review.
References
-
- Moraru M., Perez-Portilla A., Al-Akioui Sanz K., Blazquez-Moreno A., Arnaiz-Villena A., Reyburn H.T., Vilches C. FCGR Genetic Variation in Two Populations from Ecuador Highlands-Extensive Copy-Number Variation, Distinctive Distribution of Functional Polymorphisms, and a Novel, Locally Common, Chimeric FCGR3B/A (CD16B/A) Gene. Front. Immunol. 2021;12:615645. doi: 10.3389/fimmu.2021.615645. - DOI - PMC - PubMed
-
- Lalezari P., Radel E. Neutrophil-specific antigens: Immunology and clinical significance. Semin. Hematol. 1974;11:281–290. - PubMed
-
- Bux J. Granulocyte antibody-mediated neutropenias and transfusion reactions. Infusionsther. Transfusionsmed. 1999;26:152–157. doi: 10.1159/000053481. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

